Fig. 3.
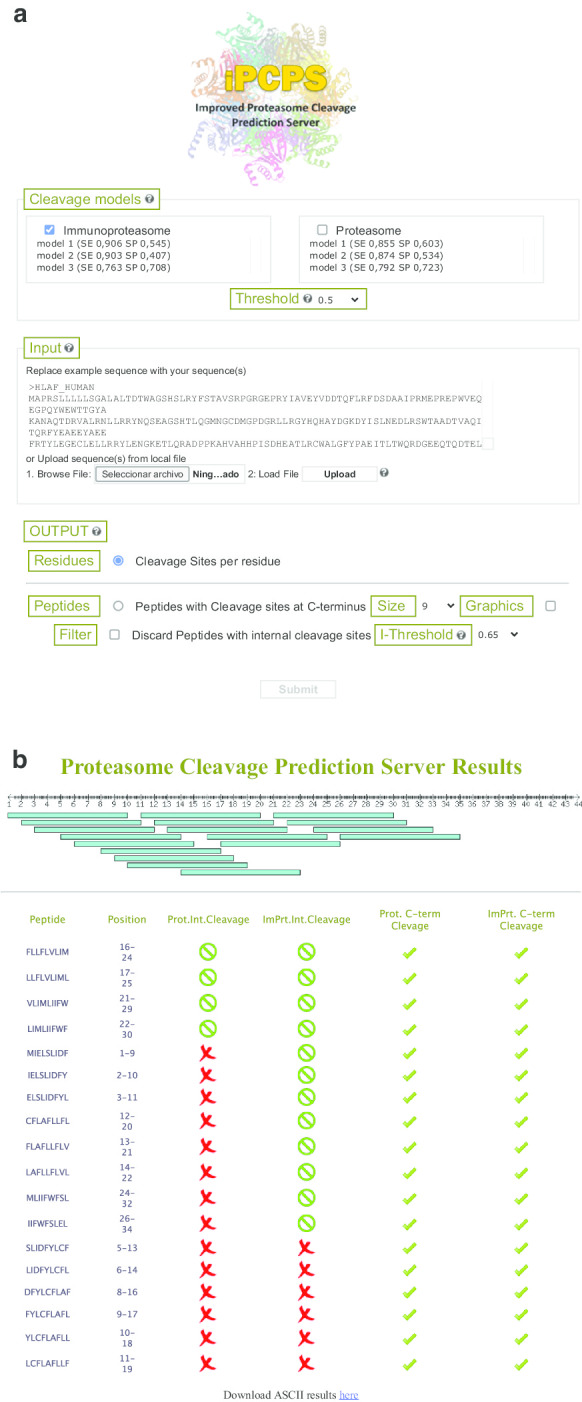
Improved Proteasome Cleavage Prediction Server (iPCPS). a Web interface of iPCPS. The interface of iPCPS is divided in three sections for intuitive use. In the first section (models), users select the immunoproteasome and/or proteasome model and the threshold. In input, users paste or upload their sequence/s in FASTA format and in output select to get cleavage sites per residue or peptides with C-terminus generated by the proteasome. Users can set the size of the peptide, discard those with internal cleavage sites and obtain a graphical display. b iPCPS output. Figure shows a representative iPCPS peptide output obtained selecting the proteasome and immunoproteasome model and graphics display. Ticks indicate that peptides have a C-terminus generated by the proteasomes and green zero symbols are for peptides without internal cleavage sites for the selected threshold. Peptides with red cross symbols do have internal cleavage sites
