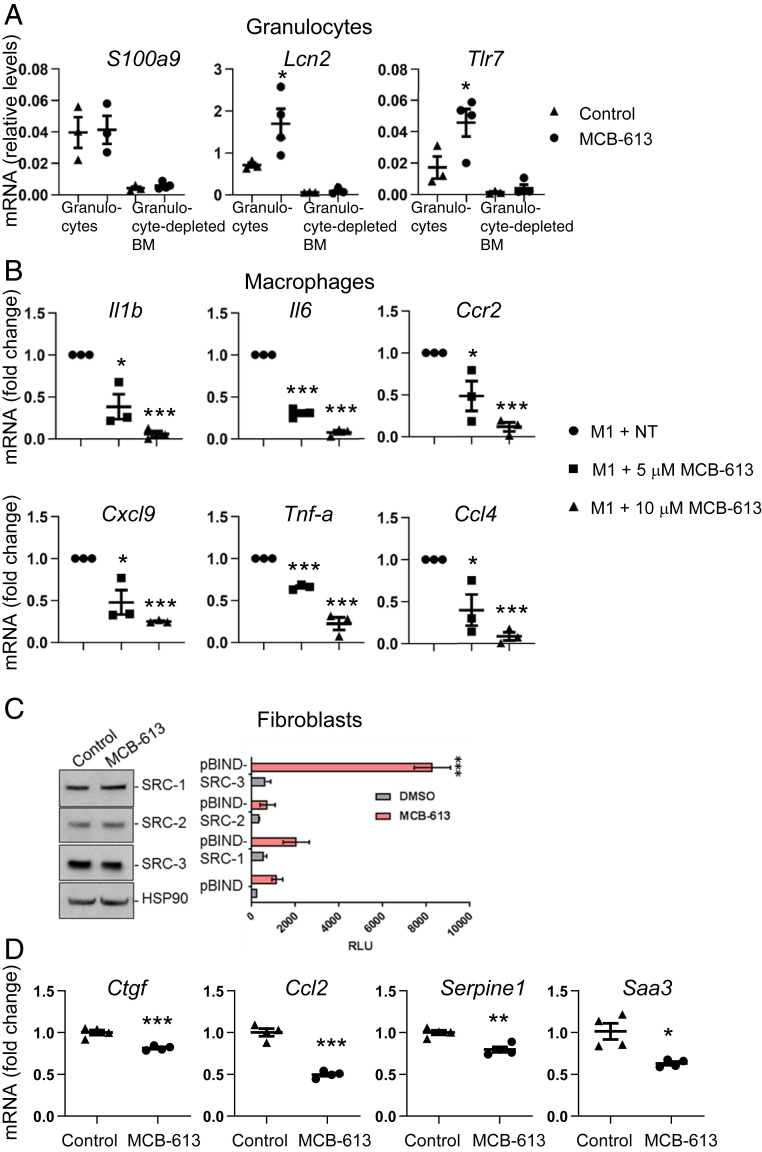
Fig. 6.
MCB-613 promotes granulocyte functions and reduces proinflammatory signaling in macrophages and fibroblasts. (A) mRNA expression in granulocytes isolated from bone marrow 24 h post-MI and MCB-613 treatment. Gene expression changes in S100a9, Tlr7, and Lcn2 were measured by qPCR and 18s RNA expression was used as a control. Values shown are mean ± SEM (n = 10 control group, n = 12 MCB-613 group); Student’s t test (Tlr7 *P = 0.01, Lcn2 *P = 0.04). (B) mRNA expression in RAW 264.7 macrophages stimulated with M1 reprogramming factors for 8 h followed by vehicle control or MCB-613 for an additional 24 h. Values shown are mean ± SEM (n = 3 control group, n = 3, 5 μM MCB-613 group, n = 3, 10 μM MCB-613 group); Student’s t test (Il1B *P = 0.01, ***P 1.3 × 10−5; Il6 ***P = 2.9 × 10−5, ***P = 3.1 × 10−6; Spp1 *P = 3.0 × 10−3, ***P = 2.4 × 10−4, Tnf-α ***P = 3.3 x 10−5, ***P = 5.0 × 10−4; Ccl4 *P = 0.03, ***P = 4.8 × 10−5; Cxcl9 *P = 0.02, ***P = 6.9 × 10−8. (C) Cardiac fibroblasts were treated with dimethylsulfoxide (DMSO) or MCB-613 for 24 h and protein was immunoblotted for SRC-1, -2, and -3. Cardiac fibroblasts were transfected with a GAL4 DNA binding site-luciferase reporter (pG5-luc) and GAL4-DNA binding domain-full-length SRC-1, -2, or -3 fusion (pBIND-SRC) or control pBIND expression vectors. Posttransfection, the cells were treated with DMSO or MCB-613 for 24 h. Values shown are mean ± SEM of a representative experiment repeated three times; Student’s t test (***P < 0.00001). (D) RT-PCR of mRNA isolated from cardiac fibroblasts 24 h after treatment with control or MCB-613. mRNA fold change for representative of three experiments, values shown are mean ± SEM n = 4 control group, n = 4, 5 μM MCB-613. Serpine1 P = 0.003, Ctgf P = 0.001, Ccl2 P = 0.0001, Saa3 P = 0.009.