Figure 1.
Striking Improvement in Model Accuracy in CASP13 Due to the Deployment of Deep Learning Methods
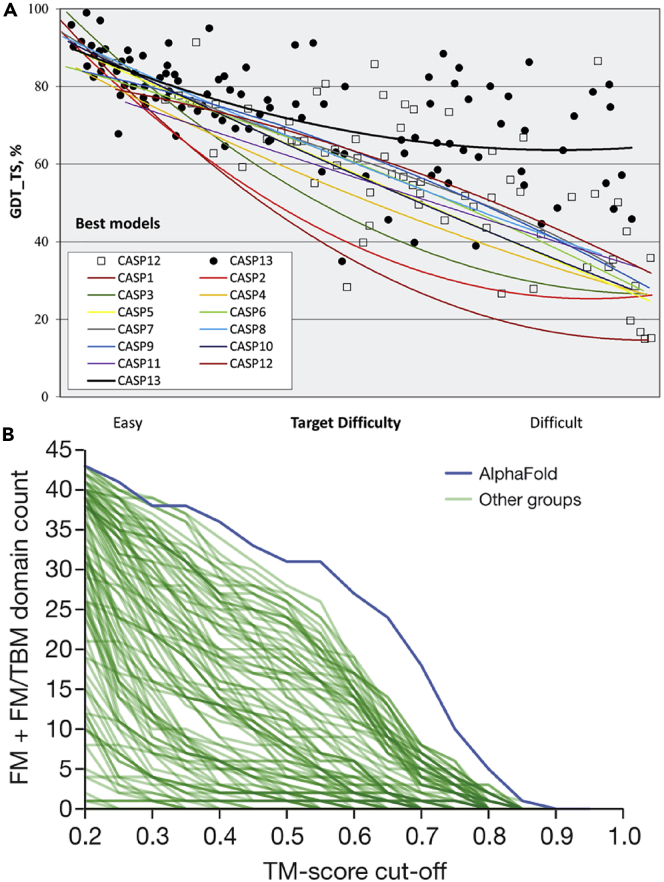
(A) Trend lines of backbone accuracy for the best models in each of the 13 CASP experiments. Individual target points are shown for the two most recent experiments. The accuracy metric, GDT_TS, is a multiscale indicator of the closeness of the Cα atoms in a model to those in the corresponding experimental structure (higher numbers are more accurate). Target difficulty is based on sequence and structure similarity to other proteins with known experimental structures (see Kryshtafovych et al.4 for details). Figure from Kryshtafovych et al. (2019).4
(B) Number of FM + FM/TBM (FM, free modeling; TBM, template-based modeling) domains (out of 43) solved to a TM score threshold for all groups in CASP.13 AlphaFold ranked first among them, showing that the progress is mainly due to the development of DL-based methods. Figure from Senior et al. (2020).26