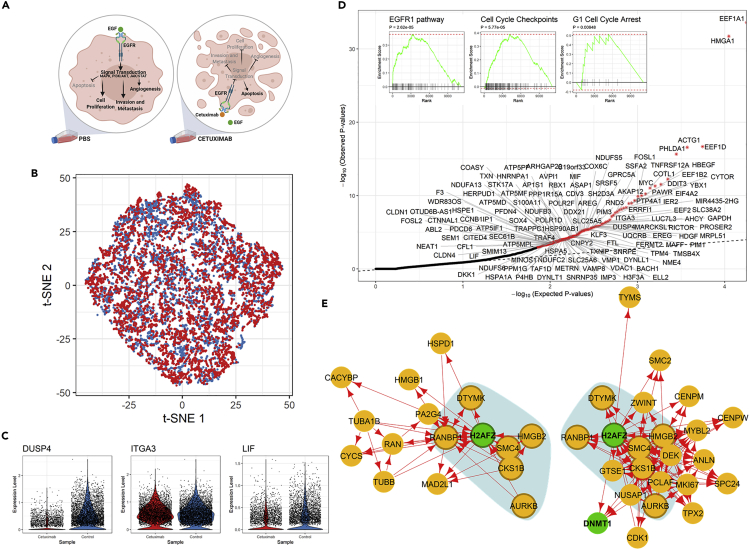
Figure 4.
Analysis of Transcriptional Responses of a Carcinoma Cell Line to Cetuximab
(A) Illustration of experimental design, including sample groups and the known mechanism of drug action, in the study of cetuximab resistance of HNSCC cell lines.48
(B) t-SNE plot of 5,217 and 4,507 HNSCC-SCC6 cells treated with cetuximab (red) and PBS (blue), respectively.
(C) Violin plots showing the log-normalized expression levels of selected differentially regulated genes in SCC6 cells with and without cetuximab treatment.
(D) Q-Q plot for observed and expected p values of the 7,503 genes tested. Genes (n = 25) with FDR <0.05 are labeled with an asterisk. Inset shows the results of the GSEA for genes ranked by their distances in manifold aligned scGRNs from young and old mice.
(E) A representative module with differentially regulated genes and corresponding subnetworks in two scGRNs. The module is enriched with differentially regulated genes and the corresponding subnetworks in two scGRNs. For illustrative purposes, the module is centered on the differentially regulated gene H2AFZ. The colors, edges, and marks are presented as in Figure 3E.