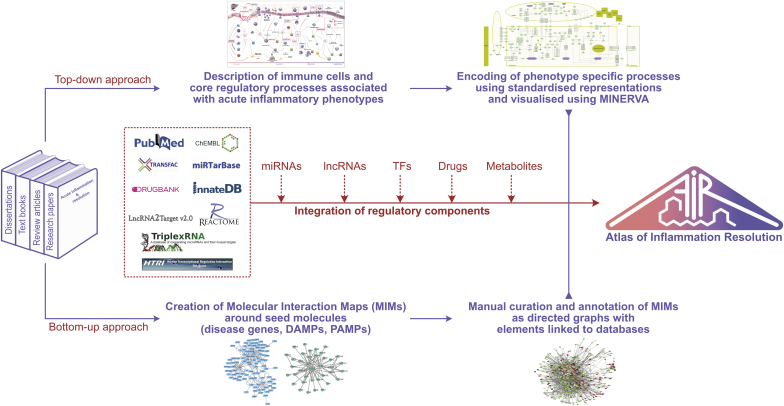
Fig. 7.
Workflow for the construction of the Atlas of Inflammation Resolution (AIR). The AIR is constructed both bottom-up and top-down. In case of the top-down approach, higher level processes, phenotypes and interplay between immune cells were identified in various stages of acute inflammation. These processes and phenotypes were extended in the form of information flow diagrams in standard SBML notations. In the bottom-up approach, first seed molecules were identified from damage-associated molecular patterns (DAMPs), Pathogen-associated molecular patterns (PAMPs) and key disease genes associated with selected clinical phenotypes of acute inflammation. Each seed molecule is then extended with the experimentally validated interacting partners. Models generated using bottom-up and top-down approaches were later merged and integrated with experimentally validated regulatory layers including transcription factors, miRNAs, lncRNAs, drugs and metabolites to prepare the AIR.