Fig. 2. Mitochondrial dysfunction and biogenesis.
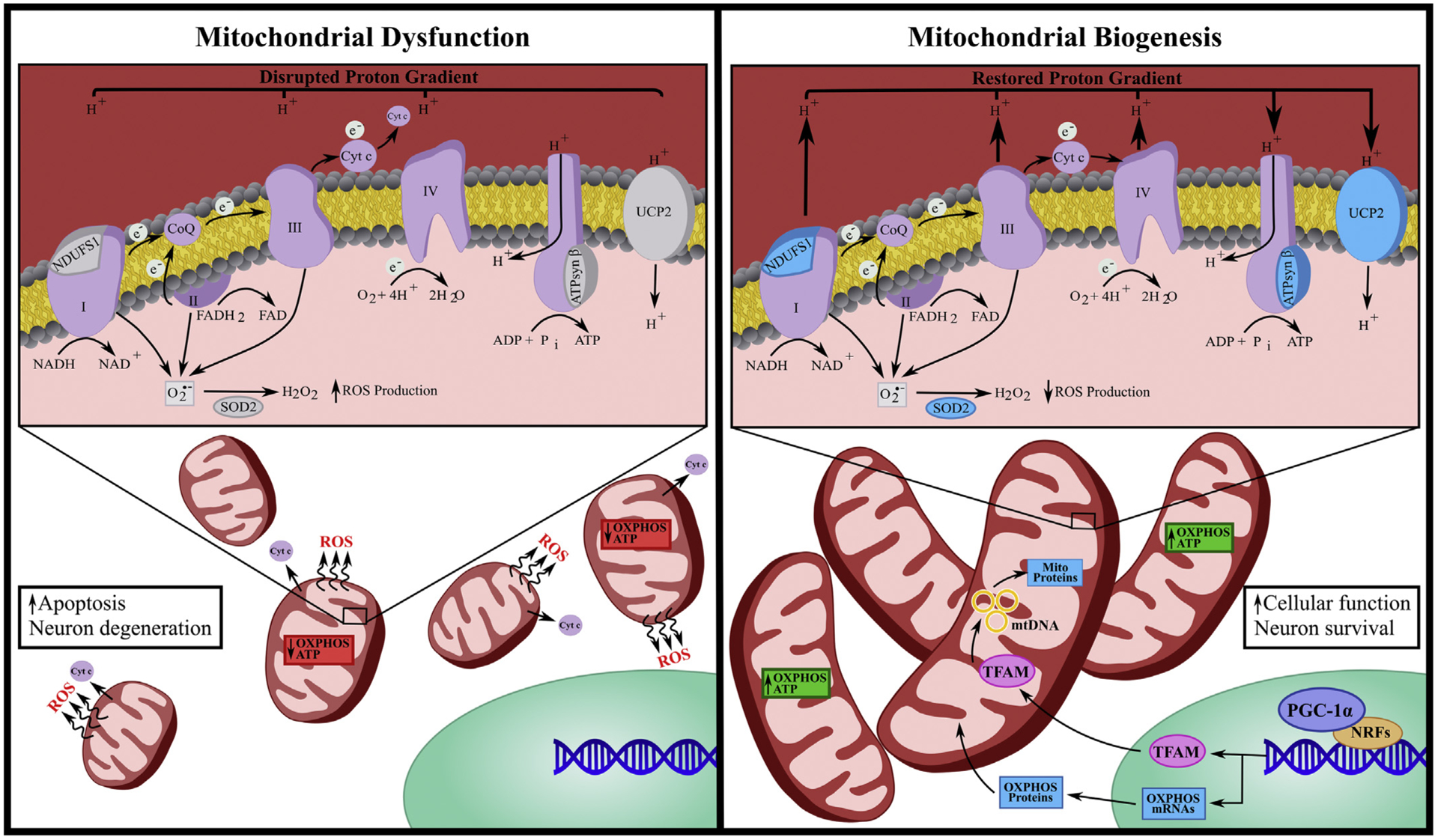
Mitochondrial dysfunction is characterized by decreased expression of oxidative phosphorylation (OXPHOS) proteins, impaired mitochondrial membrane potential, reduced ATP production as well as enhanced mitochondrial fission, reactive oxygen species (ROS) production and cytochrome c (cyt c) release. Mitochondrial dysfunction is often paired with increased apoptosis and neural degeneration. Conversely, mitochondrial biogenesis (MB) is characterized by mitochondrial fusion, reduced ROS, restoration of mitochondrial membrane potential and increased expression of OXPHOS proteins including ATP synthase β (ATPsyn β), NADH:Ubiquinone oxidoreductase core subunit 1 (NDUFS1) and uncoupling protein 2 (UCP2) as well as superoxide dismutase 2 (SOD2). MB induction often begins with activation of encoded peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) that co-activates nuclear respiratory factors (NRFs), which then promotes the transcription of several mitochondrial genes, including mitochondrial transcriptional factor a (TFAM). Once translated, TFAM translocates to the mitochondrial matrix and stimulates mtDNA replication and mitochondrial gene expression. MB is associated with enhanced cellular function and neuronal survival.
