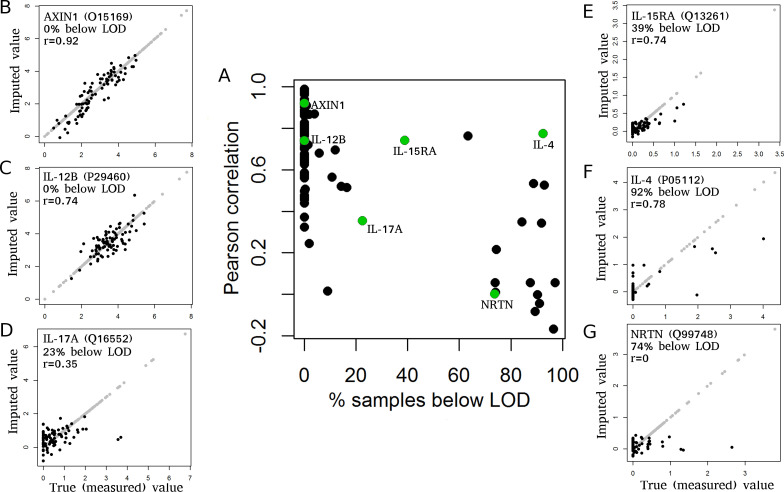
Fig 5. GSimp imputation vs. remeasurement.
(A) Pearson correlation between GSimp-imputed and remeasured protein expression values (y-axis) versus fraction of samples below LOD (x-axis) for 91 proteins. The results were overall similar to those based on missForest (Fig 2). (B-G) Scatterplots of remeasured (x-axis) versus imputed (y-axis) values for selected proteins. Protein names, fractions of samples below LOD and Pearson correlation value (r) are provided for each panel. Each dot represents one sample. Black dots represent the imputed and remeasured values. Light gray dots represent all values that did not have to be imputed and are visualized as reference. Scatterplots for all 91 proteins are provided in S9 Fig. The scatterplot of IL-12B (C) revealed no reduction in variance for imputed values, in contrast to the missForest result (Fig 2C).