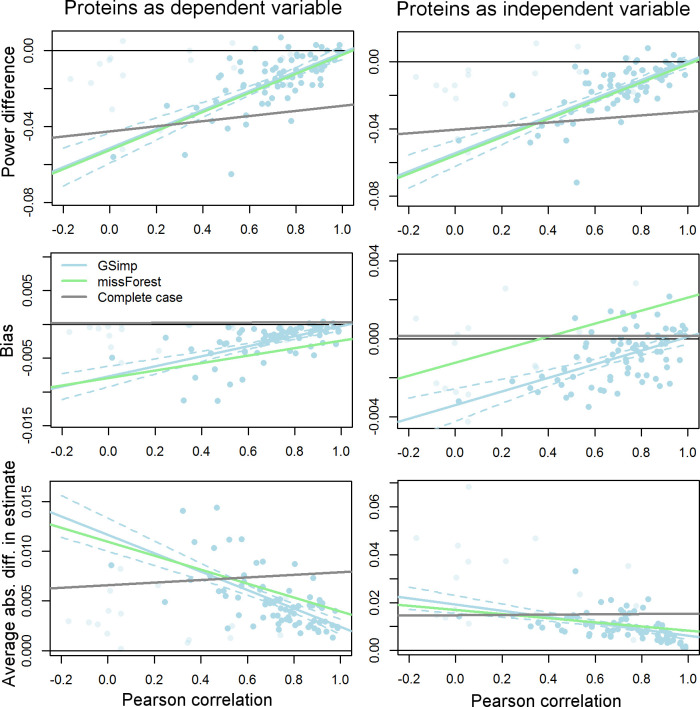
Fig 6. Effect of imputation on downstream analyses.
Evaluation of imputation effects on downstream analyses utilizing the proteome data as dependent (left) or independent (right) variable in univariate regression models. The power, bias, and average absolute difference in univariate regression estimates to the complete dataset are shown for GSimp-imputed (light blue dots and lines), missForest imputed (green line), or non-imputed, i.e. complete case, (gray line) data. The simulation utilized all 10 chips, and a beta value of 0.01. For GSimp and missForest, the power increases, and average absolute difference decreases with increasing correlation between imputed and remeasured data. The bias decreases with increasing imputation accuracy when using protein data as dependent variable and crosses zero when using protein data as independent variable. An empirical correlation cutoff of 0.4 (power) or 0.5 (average absolute difference) is observed above which imputation is beneficial compared to no imputation (complete case analysis). Imputation with GSimp results in slightly lower bias and lower absolute average difference compared to missForest for proteins with high imputation accuracy. Proteins with more than 25% of values below LOD are colored transparently and were excluded for calculation of regression lines.