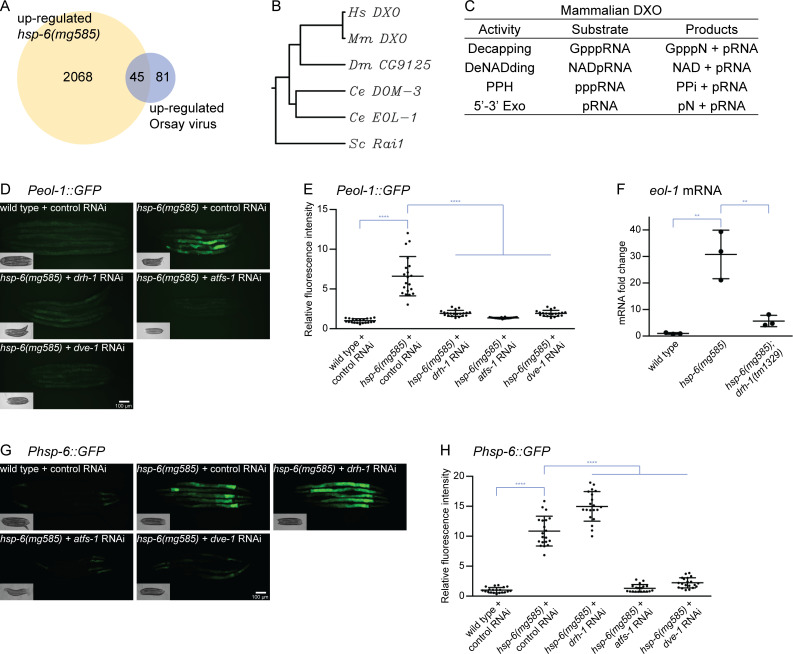
Fig 2. Increased eol-1 expression depends on DRH-1.
(A) Venn diagram of genes up-regulated in hsp-6(mg585) and Orsay virus infection. (B) Phylogenetic tree of EOL-1/DXO. EOL-1 is conserved from yeast to mammals. (C) The enzyme activities of human DXO. Mammalian DXO modifies the 5′ end of mRNAs: decapping, deNADing, pyrophosphohydrolase and 5′-3′ exonuclease. (D and E) The induction of Peol-1::GFP transcriptional fusion reporter requires drh-1 and UPRmt. The Peol-1::GFP is strongly induced by hsp-6(mg585) mitochondrial mutant, and this induction is abrogated by drh-1 RNAi as well as RNAi of genes involved in UPRmt (atfs-1 and dve-1). Animals were imaged in (D) and the fluorescence was quantified in (E). **** denotes p < 0.0001. (F) The hsp-6(mg585) mutant causes increased mRNA level of eol-1 in a drh-1-dependent manner. RT-qPCR assays showed the mRNA level of eol-1 was induced by hsp-6(mg585) mutant, and this induction was abolished in hsp-6(mg585); drh-1(tm1329) double mutant. ** denotes p < 0.01. (G and H) DRH-1 does not contribute to UPRmt. The benchmark reporter of UPRmt Phsp-6::GFP is induced by the hsp-6(mg585) mutant. And the induction is suppressed by RNAi of atfs-1 or dev-1, but not drh-1. Animals were imaged in (G) and the fluorescence was quantified in (H). **** denotes p < 0.0001. The underlying numerical data can be found in S1 data. Ce: Caenorhabditis elegans; Dm: Drosophila melanogaster; Hs: Homo sapiens; Mm: Mus musculus; RT-qPCR, quantitative reverse transcription PCR; Sc: Saccharomyces cerevisiae. UPRmt, mitochondrial unfolded protein response.