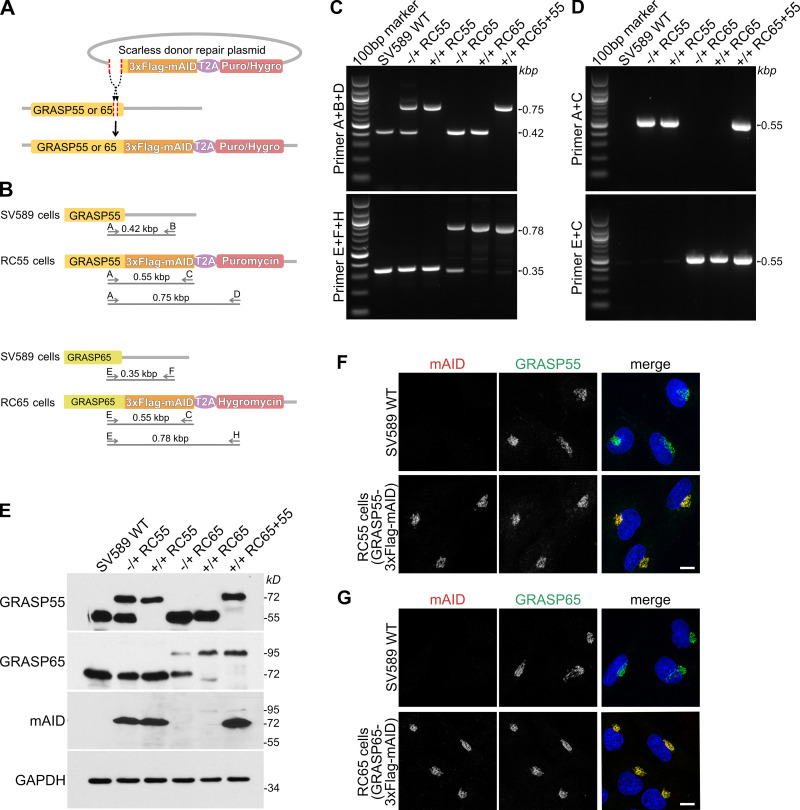
Figure 1.
Generation of cell lines expressing endogenous GRASP55 and/or 65 tagged with mAID. (A) Scheme for tagging the endogenous loci of GRASP55 or 65 with 3xFlag and mAID. The DNA sequence encoding 3xFlag-mAID separated by a self-cleaving T2A peptide from the resistance gene (puromycin [Puro] or hygromycin [Hygro]) was inserted by CRISPR-Cas9 gene editing in front of the stop codon of GRASP55 or 65. The amino acids between the insert location and the stop codon were scarlessly repaired by adding the sequence in front of 3xFlag-mAID of the repair donor plasmid. (B) Scheme of the primer sets used for genotyping gene-edited cell lines. The cell line with GRASP55 scarlessly tagged with 3xFlag-mAID and puromycin is referred to as RC55, GRASP65 scarlessly tagged with 3xFlag-mAID and hygromycin as RC65, and both GRASP55 and 65 tagged with 3xFlag-mAID and indicated resistance genes as RC65+55. (C and D) Genomic PCR to genotype heterozygous (−/+) and homozygous (+/+) RC55, RC65, and RC65+55 cell lines for GRASP55 tagged with 3xFlag-mAID or GRASP65-3xFlag-mAID. Parental SV589 cells are shown as negative control (WT). (E) Immunoblot analysis of whole cell lysates of heterozygous (−/+) and homozygous (+/+) RC55, RC65, and RC65+55 cells using antibodies against GRASP55, GRASP65, the degron mAID tag, and GAPDH. (F) SV589 cells and RC55 cells expressing GRASP55-3xFlag-mAID were immunostained for mAID (red) and GRASP55 (green) and labeled for DNA (blue). Scale bar, 10 μm. (G) SV589 cells and RC65 cells stably expressing GRASP65-3xFlag-mAID were immunostained for mAID (red) and GRASP65 (green) and labeled for DNA (blue). Scale bar, 10 μm.