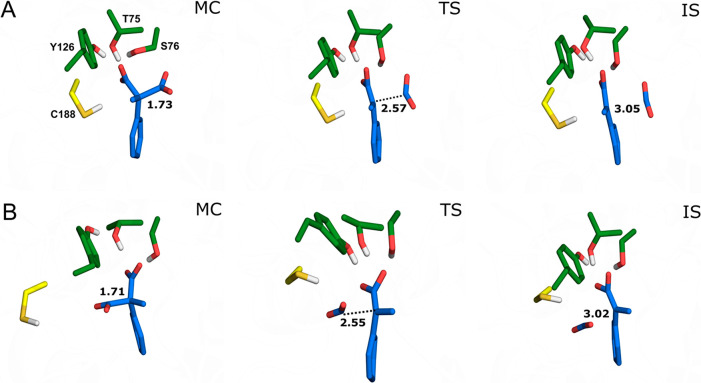
Figure 3.
Representative structures of the Michaelis complexes (MC), transition states (TS), and intermediate states (IS), for cleavage of (A) the pro-(R) and (B) the pro-(S) carboxylate groups of compound 1a by wild-type AMDase, as obtained from EVB simulations of these reactions. For the full reaction mechanism, see Scheme 1. The structures shown here are the centroids of the top ranked cluster obtained from clustering on RMSD, performed as described in the SI. The labeled C–C distances are averages at each stationary point over all trajectories (see Table S1). Corresponding representative structures of key stationary points during simulations of the wild-type AMDase catalyzed decarboxylation of compounds 1b to 1e can be found in Figures S8–S11. The color-coding of key residues follows that of Figure 1A.