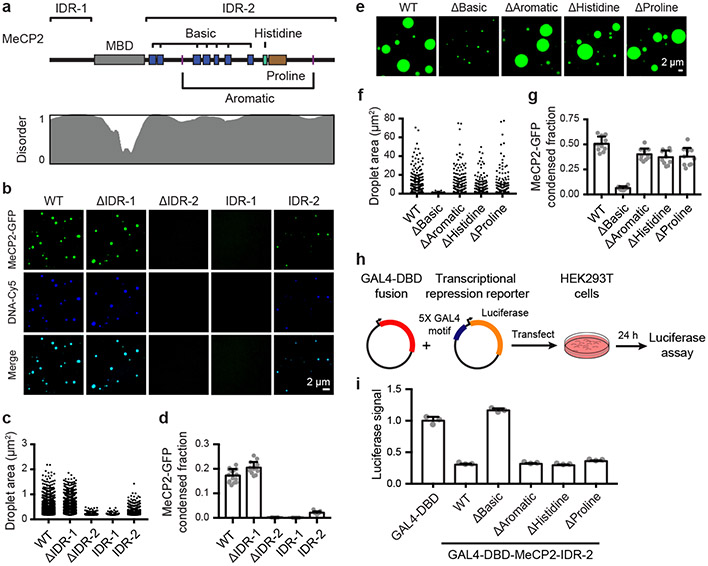
Figure 2. MeCP2 features that contribute to condensate formation.
a. Schematic of MeCP2 protein indicating the MBD, IDR-1, IDR-2, and sequence features within IDR-2 previously implicated in condensate formation for other proteins. Contribution of IDR-2 sequence features to condensate formation was examined using deletion mutants that remove the basic patches (ΔBasic), aromatic residues (ΔAromatic), histidine-rich patch (ΔHistidine), and proline-rich patch (ΔProline). Predicted protein disorder is displayed below.
b. Droplet experiments examining ability of MeCP2 deletion mutants to form droplets with DNA. MeCP2-GFP deletion mutants at 2 μM were mixed with 40 nM DNA in droplet formation buffers with 100 mM NaCl.
c. Droplet areas for experiments in Fig. 2b. Fields per condition n=15.
d. MeCP2-GFP condensed fraction for experiments in Fig. 2b. Mean±SD. Fields per condition n=15.
e. Droplet experiments examining ability of MeCP2 IDR-2 sequence feature deletion mutants to form droplets. MeCP2-GFP IDR-2 sequence feature deletion mutants at 10 μM were added to droplet formation buffers with 150 mM NaCl and 10% PEG-8000.
f. Droplet areas for experiments in Fig. 2e. Fields per condition n=10.
g. MeCP2-GFP condensed fraction for experiments in Fig. 2e. Mean±SD. Fields per condition n=10.
h. Schematic of transcriptional repression reporter assay used to examine the ability of MeCP2 IDR-2 sequence features to contribute to transcriptional repression.
i. Normalized luciferase signals for reporter assay examining ability of MeCP2 IDR-2 sequence features to contribute to transcriptional repression. Luciferase signal was normalized to GAL4-DBD alone. Mean±SD, n=3 biologically independent samples per condition.