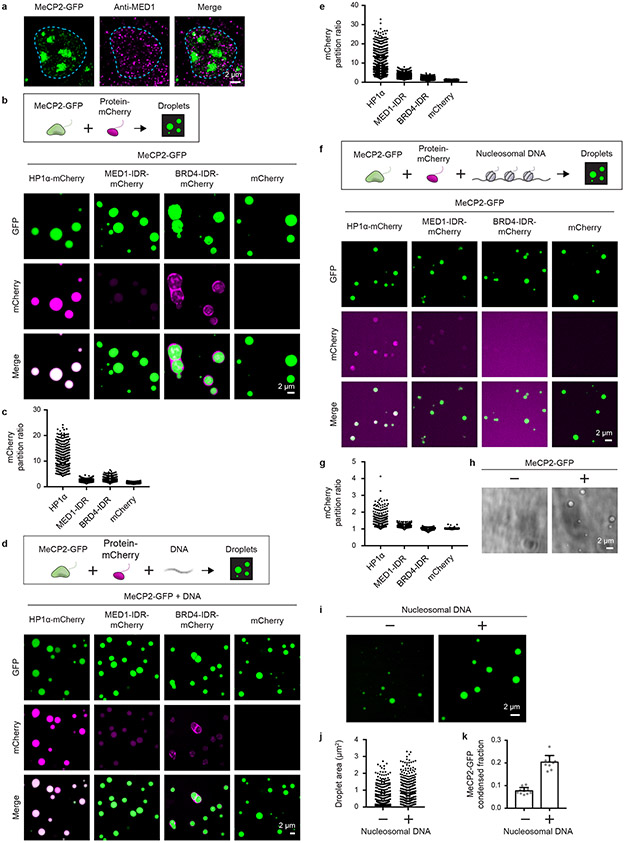
Extended Data Figure 4. MeCP2 condensates preferentially concentrate HP1α compared to components of transcriptional condensates.
a. Immunofluorescence images of heterochromatin condensates (MeCP2-GFP) and transcriptional condensates (Anti-MED1) in mESCs.
b. Droplet experiments examining ability of MeCP2 condensates to preferentially concentrate HP1α compared to components of transcriptional condensates. MeCP2-GFP at 7.5 μM was mixed with HP1α-mCherry, MED1-IDR-mCherry, BRD4-IDR-mCherry, or mCherry at 7.5 μM in droplet formation buffers with 150 mM NaCl and 10% PEG-8000.
c. mCherry partition ratios in MeCP2-GFP droplets for experiments in Extended Data Fig. 4b. Fields per condition n=15.
d. Droplet experiments with naked DNA examining ability of MeCP2 condensates to preferentially concentrate HP1α compared to components of transcriptional condensates. Conditions same as in Extended Data Fig. 4b, but with addition of 160 nM DNA.
e. mCherry partition ratios in MeCP2-GFP droplets for experiments in Extended Data Fig. 4d. Fields per condition n=15.
f. Droplet experiments with nucleosomal DNA examining ability of MeCP2 condensates to preferentially concentrate HP1α compared to components of transcriptional condensates. MeCP2-GFP at 5 μM was mixed with HP1α-mCherry, MED1-IDR-mCherry, BRD4-IDR-mCherry, or mCherry at 5 μM and 6 nM poly-nucleosomes in droplet formation buffers with 100 mM NaCl and 3 mM MgCl2.
g. mCherry partition ratios in MeCP2-GFP droplets for experiments in Extended Data Fig. 4f. Fields per condition n=10.
h. Brightfield images examining droplet formation with nucleosomal DNA alone and with MeCP2. Poly-nucleosomes at 6 nM were mixed with 5 μM MeCP2-GFP or no MeCP2-GFP in droplet formation buffers with 100 mM NaCl and 3 mM MgCl2.
i. Droplet experiments examining MeCP2 droplet formation with nucleosomal DNA. Conditions same as in Extended Data Fig. 4h.
j. Droplet areas for experiments in Extended Data Fig. 4i. Fields per condition n=10.
k. MeCP2-GFP condensed fraction for experiments in Extended Data Fig. 4i. Mean±SD. Fields per condition n=10.