Fig. 5. Sequence and structure-based pathogenicity predictions of NLR inflammasome variants.
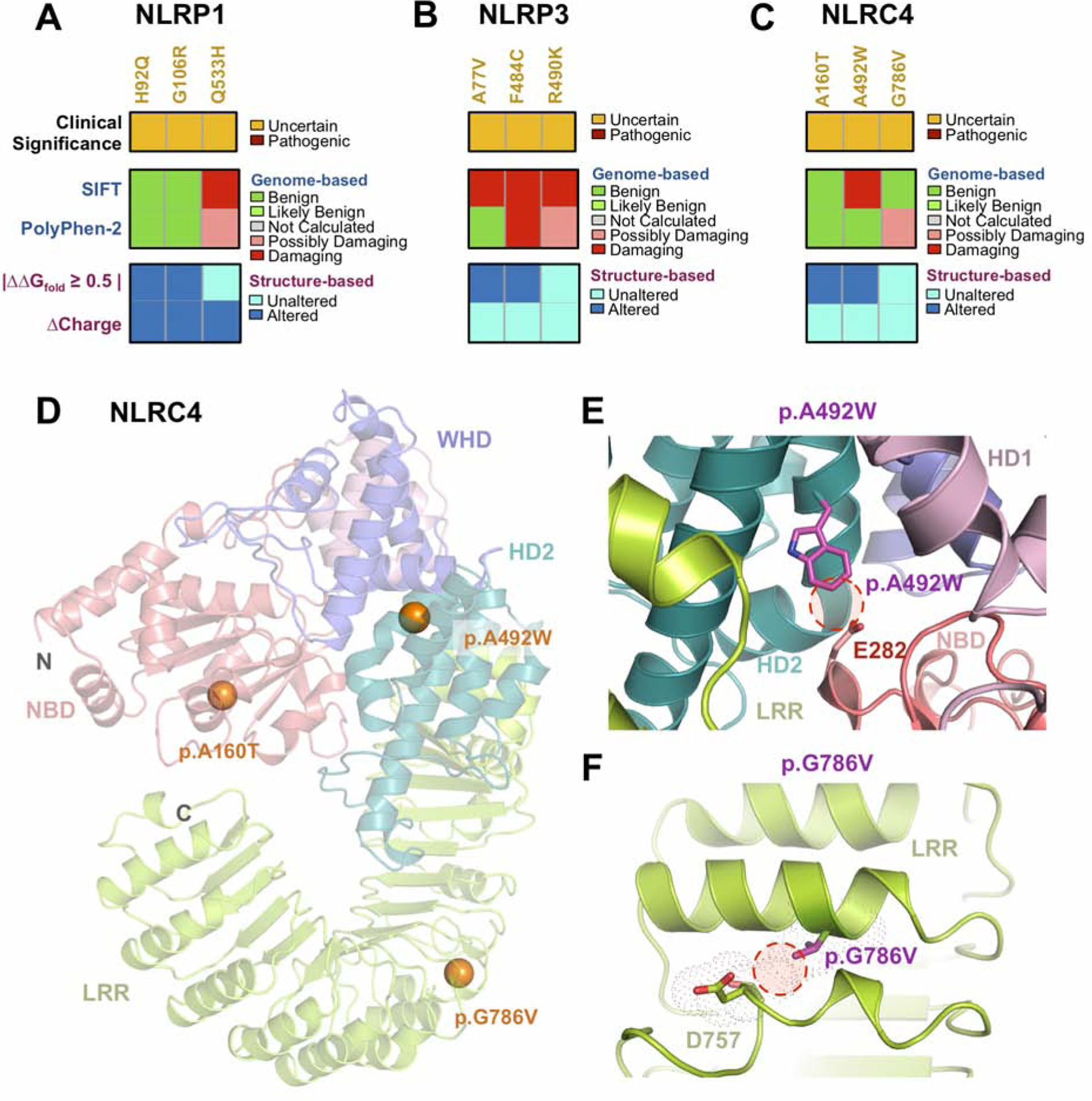
Sequence- and structure-based pathogenicity predictions for (A) NLRP1, (B) NLRP3, and (C) NLRC4 variants identified in uveitis patients. Variants are annotated by their clinical significance (uncertain or pathogenic). Sequence-based predictors (SIFT, PolyPhen-2) provide predictions of degree of damage for individual variants, while structure-based predictors provide information on specific mechanistic alterations. (D) Ribbon trace diagram of the murine NLRC4 structure (PDB 4KXF) highlighting the different functional domains. Variants identified in the WES screen are represented as spheres and are colored according to their clinical significance – orange, unknown; red – confirmed pathogenic. (E) Close up view of the p.A492W variant in the HD2 domain. Introduction of a Trp residue results in steric clashes between residues of the HD2 and NBD domain. (F) Close up view of the p.G786V variant in the LRR domain. This substitution likely introduces steric clashes in the neighboring α-helix.
