Figure 1. PPAR structure and mechanism of action.
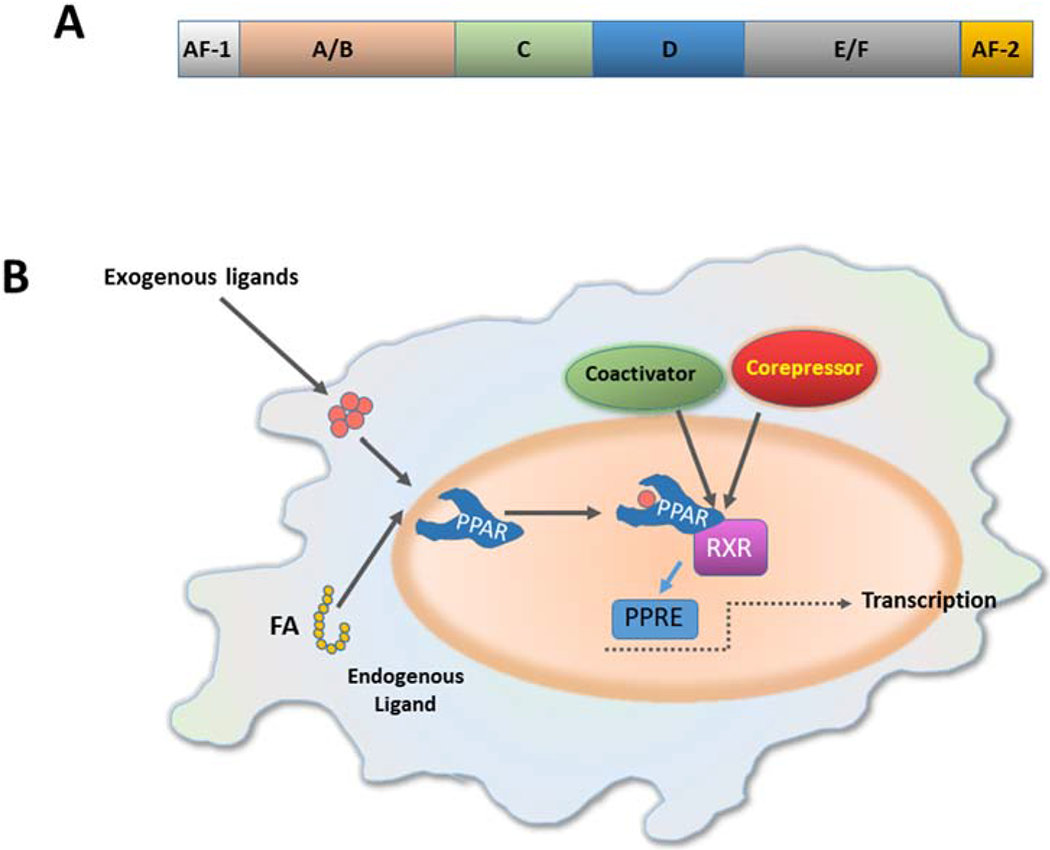
(A) All PPARs share the basic structural of the most nuclear receptors, consisting of four functional domains named A/B, C, D and E/F. The N-terminal A/B domain contains a ligand-independent activation function 1 (AF-1). The central DNA binding domain (DBD) or C domain is a conserved domain, composed of two zing fingers, and is responsible for the binding of PPAR to the peroxisome proliferator response element (PPRE) in the promoter of target genes. The D domain is a docking site for various cofactors. The E domain or ligand-binding domain (LBD) binds a variety of endogenous or exogenous lipophilic ligands and provides ligand specificity. Recruitment of PPAR cofactors that participate in the transcription process is mediated by the ligand-dependent activation function-2 (AF-2), located in the E/F domain. (B) Ligand binding promotes conformational changes that enable the interaction with co-activator complexes. The full transcriptional activity of PPARs requires binding of cognate lipid ligands, heterodimerization with another nuclear receptor (retinoid-X receptor, RXR), interaction with a number of transcriptional coactivators, and binding of the complex to PPAR response elements (PPREs) in the promotor of target genes. (FA, fatty acid)
