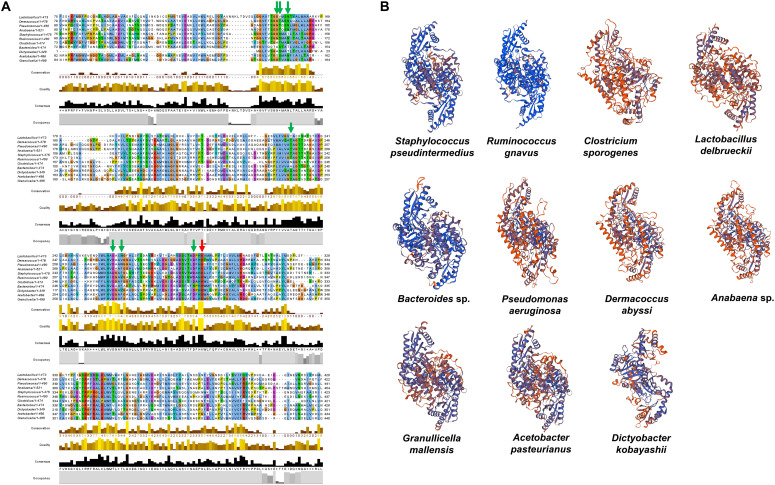
FIGURE 6.
SadA homologs of several bacterial species from different phyla. (A) We aligned multiple sequences of SadA homologs of different species from different phyla. Jalview 2.11 was used for visualization (Letunic and Bork., 2019). The PLP-binding motifs are marked with green arrows and the active site lysine with a red arrow. (B) We also predicted the 3D structure of SadA homologs using SWISS-MODEL (31). Staphylococcus pseudintermedius, Clostridium sporogenes, Lactobacillus delbrueckii, and Ruminococcus gnavus represent Firmicutes; Dermacoccus abyssi represents Actinobacteria; Pseudomonas aeruginosa and Acetobacter pasteurianus represent Proteobacteria; Anabaena sp. represents Cyanobacteria; Bacteroides sp. represents Bacteroidetes; Dictyobacter kobayashii represents Chloroflexi; and Granulicella mallensis represents Acidobacteria. Accession numbers of the SadA homologs used for the alignment: Staphylococcus pseudintermedius WP_014612792, Ruminococcus gnavus A7B1V0.1, Clostridium sporogenes WP_058007894.1, Lactobacillus delbrueckii WP_130183735.1, Pseudomonas aeruginosa MXH35303.1, Dermacoccus abyssi QEH92696.1, Acetobacter pasteurianus GAB29948.1, Bacteroides sp. KPL16553.1, Granulicella mallensis WP_014266912.1, Dictyobacter kobayashii GCE20849.1, and Anabaena sp. HFS08142.1.