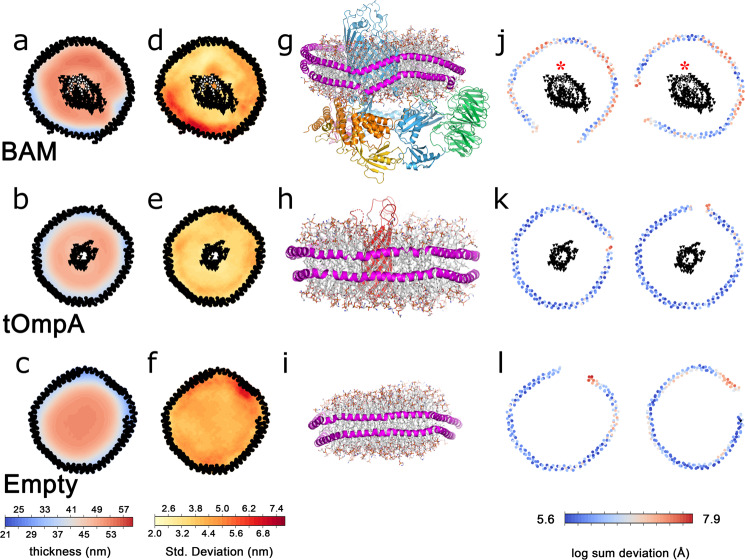
Fig. 4. Molecular dynamics simulations of protein-containing and empty lipid nanodiscs.
Mean thickness of lipid bilayers over the course of a 1-µs simulation for BAM-containing (a), tOmpA-containing (b) or empty (c) MSP1D1 nanodiscs. The standard deviation of bilayer thickness over the 1-μs simulation for BAM-containing (d), tOmpA-containing (e) or empty (f) MSP1D1 nanodiscs. Residues in the lateral gate are indicated in white. The 3D structures of the final frame of each simulation, coloured as in Fig. 1, with the MSP in magenta for BAM-containing (g), tOmpA-containing (h) or empty (i) MSP1D1 nanodiscs. Deviation from planarity for the upper (left) and lower (right) MSP in BAM-containing (j), tOmpA-containing (k) or empty (l) MSP1D1 nanodiscs. In a, b, d, e, j and k the mean positions of the α-carbons of the MSPs and β-barrels of the inserted proteins are represented by black dots. Strands β1 and β16, which make up the lateral gate of BamA, are indicated with a red asterisk in (j).