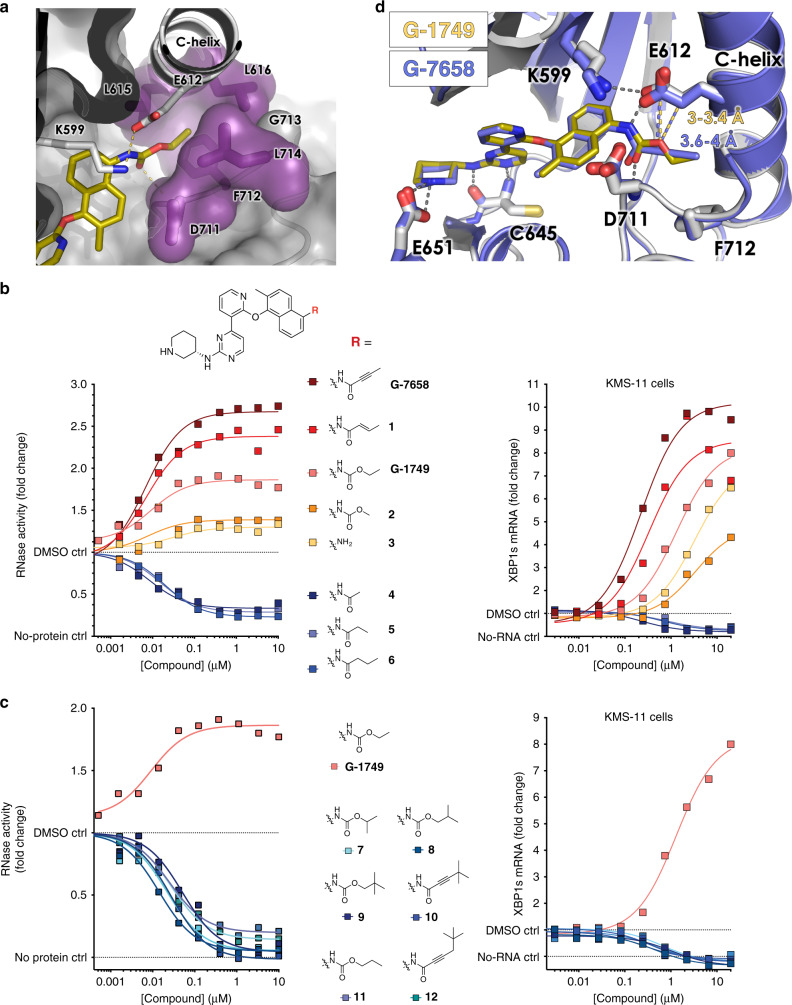
Fig. 3. The nature of the tail substituent determines the degree of RNase activation.
a Co-crystal structure of G-1749 (golden sticks) in complex with unphosphorylated IRE1 (gray cartoon). Details of the front pocket where G-1749 ethyl carbamate tail binds are highlighted as magenta sticks and surface. b, c Left, dose-response curves in biochemical RNA cleavage assay; right, XBP1s mRNA levels measured by bDNA assay after treatment with compounds for 4 h, for indicated derivatives of G-1749. In all experiments background from the no-protein and no-RNA controls were subtracted from signal before calculating fold change. Source data are provided as a Source Data file. Data are presented as the mean for measurements from two independent experiments (n = 2). d Co-crystal structure of G-7658 (purple sticks) in complex with unphosphorylated IRE1 (purple cartoon) overlayed on the co-crystal structure of G-1749 (golden sticks) in complex with unphosphorylated IRE1 (gray cartoon). Conserved hydrogen bonds are shown as gray dashes. The distances between E612 and the oxygen of G-1749 carbamate tail or the alkynyl carbon of G-7658 tail are highlighted as golden or purple dashes, respectively, and the measured distance is reported in the corresponding color.