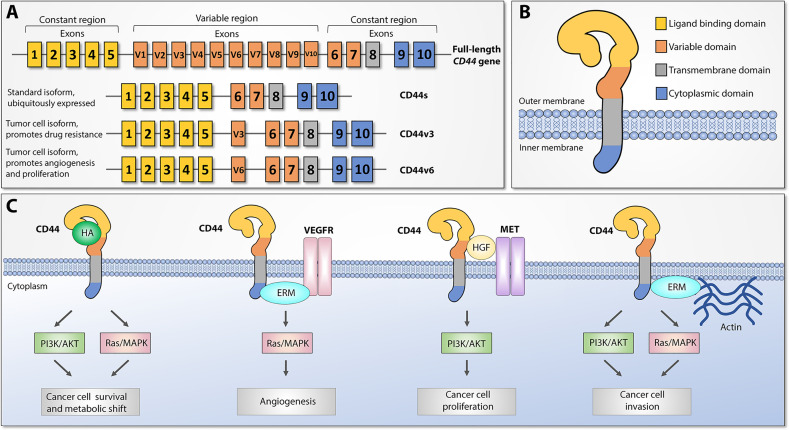
Figure 1.
CD44 structure and downstream signaling pathways, adapted from (12). (A) Top: CD44 gene structure. CD44 full-length pre-mRNA consists of 20 (mice) or 19 (human) exons, the first and last 5 of which are constant and 9-10 exons in the middle are variable (v) exons regulated by alternative splicing. Bottom: standard (CD44s) and most widely studied cancer-associated alternatively spliced variant isoforms (CD44v3 and CD44v6). Exon coloring parallels corresponding protein domains. (B) CD44 protein structure. Four main regions of the CD44 protein are presented with exon matching colors: constant extracellular ligand binding domain, variable extracellular domain, constant transmembrane domain, and cytoplasmic domain. (C) Main CD44-mediated downstream signaling pathways. Canonical CD44 activation relies on extracellular ligand stimulation, such as HA and subsequent PI3K and MAPK pathway activation, which leads to cancer cell metabolic shift and resistance to apoptotic stimuli. Via intracellular ERM protein recruitment, the cytoplasmic tail of CD44 can either interact with VEGFR and support tumor angiogenesis or promote cytoskeletal changes and promote cancer cell invasion. Additionally, CD44 may act as a coreceptor for several receptor tyrosine kinases, such as Met, to facilitate cancer progression. ERM: ezrin, radixin, and moesin. VEGFR: Vascular endothelial growth factor receptor.