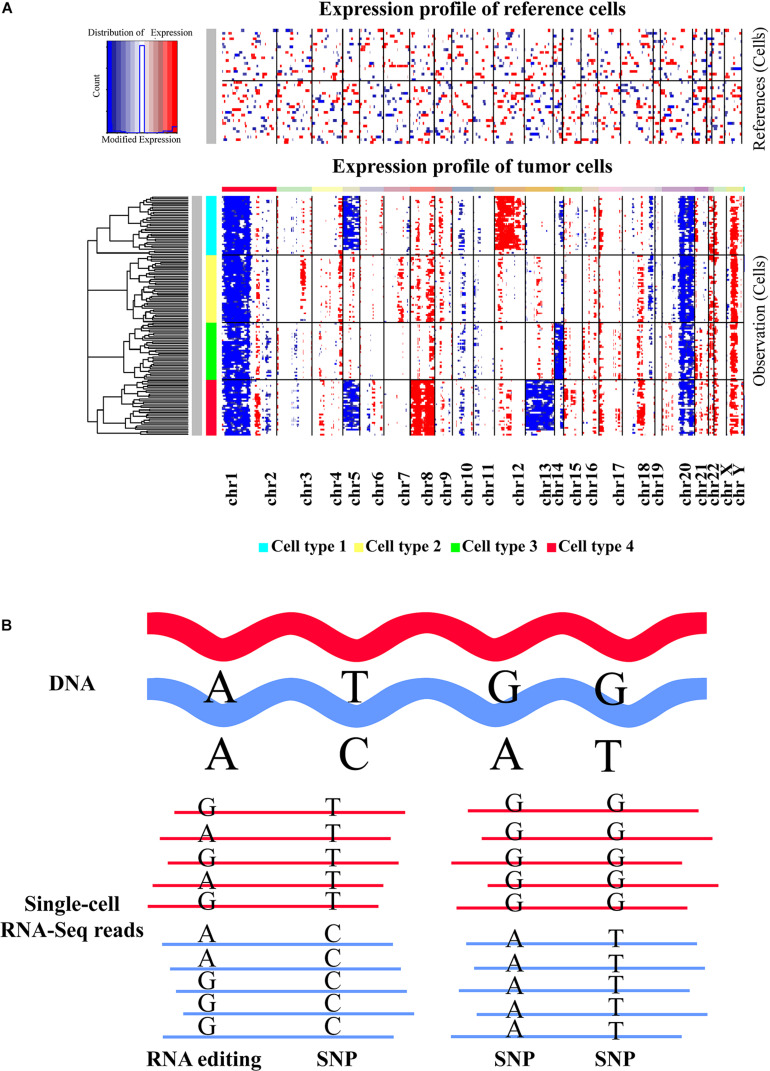
FIGURE 3.
Inference of large-scale copy number alterations and single nucleotide changes based on scRNA-seq data. (A) Representative heatmap displaying the large-scale copy number variations (CNVs) identified in different cell types with scRNA-seq data. Top panel shows that no significant large-scale CNVs were identified in reference normal cells, whereas chromosomal-scale deletions (blue) and gains (red) were observed for several chromosomes in different cell subtypes of tumor cells (second panel). The heatmap was created by inferCNV. (B) Graphic view of single nucleotide variations and RNA editing events. The reads of scRNA-seq data generated from full-length transcript sequencing protocols are mapped to the reference genome first. Then specific SNV calling tools or RNA-editing detection approaches can be applied to determine the SNVs or RNA-editing events based on the alignment result. Both SNV and RNA editing identifications are mainly suitable for the scRNA-seq methods that can generate full-length transcripts. Moreover, sequencing depth could be an important factor influencing the detection accuracy.