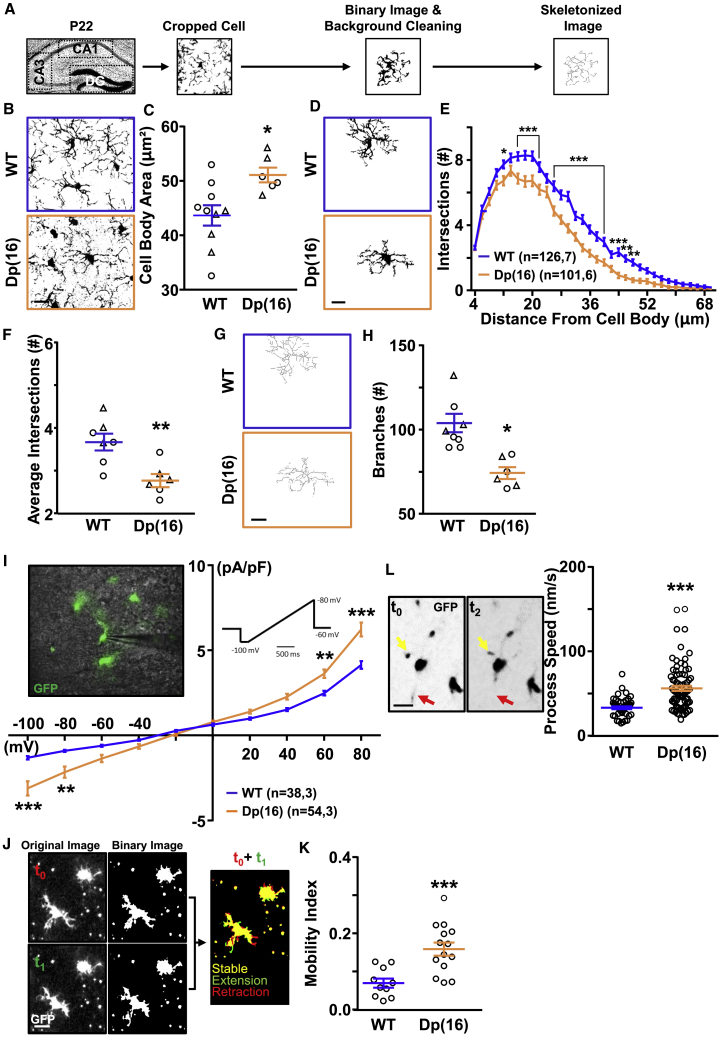
Figure 1.
Microglial Morphology and Activity Are Altered in Dp(16) Hippocampi
(A) Experimental protocol.
(B) Iba1-stained hippocampal slices from P22 WT and Dp(16) animals. Scale bar: 10 μm.
(C) Quantification of the microglial cell body area. Bars represent the average of microglial cell body areas in all analyzed animals ± SEM, and symbols (circles, males; triangles, females) represent data points for each animal (38–73 cells/animal; 1 slice per animal). ∗p < 0.05; unpaired two-tailed Student’s t test; t = 2.8; df = 14.
(D) Binary images of selected cells from the fields in (B). Scale bar: 10 μm.
(E) Sholl analysis of microglial cells. Data are expressed as average number of intersections at each distance from cell bodies of all analyzed cells ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; two-way ANOVA; FStrain (33, 7,650) = 317.5; p < 0.001; Holm-Sidak post hoc test. In parenthesis: analyzed cells, animals (1 slice per animal).
(F) Quantification of the average number of intersections from the Sholl analysis. Bars represent the average number of intersections in all analyzed animals ± SEM, and symbols (circles, males; triangles, females) represent data points for each animal (12–21 cells/animal; 1 slice per animal). ∗∗p < 0.01; unpaired two-tailed Student’s t test; t = 3.47; df = 11.
(G) Representative skeleton images of binary images. Scale bar: 10 μm.
(H) Quantification of the number of branches per microglial cell. Bars represent the average number of branches per microglial cell ± SEM in all analyzed animals; symbols (circles, males; triangles, females) represent data points for each animal (12–21 cells/animal; 1 slice per animal). ∗p < 0.05; unpaired two-tailed Student’s t test; t = 4.03; df = 11.
(I) Voltage-activated current profile in GFP+ microglia of acute hippocampal slices from WTCX3CR1-GFP and Dp(16)CX3CR1-GFP animals (left inset). Voltage currents were analyzed using ramps ranging from −100 to 80 mV (right inset). Data are expressed as average current for each mV step of all analyzed cells ± SEM. ∗∗p < 0.01; ∗∗∗p < 0.001; two-way repeated measures (RM)ANOVA; FInteraction (9, 900) = 13.35; p < 0.001; Holm-Sidak post hoc test. In parenthesis: analyzed cells, animals.
(J) Confocal and binary example images of a P22 WTCX3CR1-GFP hippocampal slice in time-lapse experiments at different time points. The arrow points to the merge of the t0 and the t1 image used for quantifying pixel intensity changing over time. Scale bar: 20 μm.
(K) Quantification of the mobility index of microglial cells. Bars represent the average mobility index in all analyzed fields ± SEM, and circles represent data points for each field (3 animals per genotype). ∗∗∗p < 0.001; unpaired two-tailed Student’s t test; t = 3.98; df = 22.
(L) Representative images of GFP+ moving cells (4.5 min apart). Scale bar: 20 μm (left). Quantification of process speed of moving microglial cells is shown (right). Bars represent the average process speed in all analyzed cells ± SEM, and circles represent data points for each cell (3 animals per genotype). ∗∗∗p < 0.001; two-tailed Mann-Whitney test; U = 701.5.