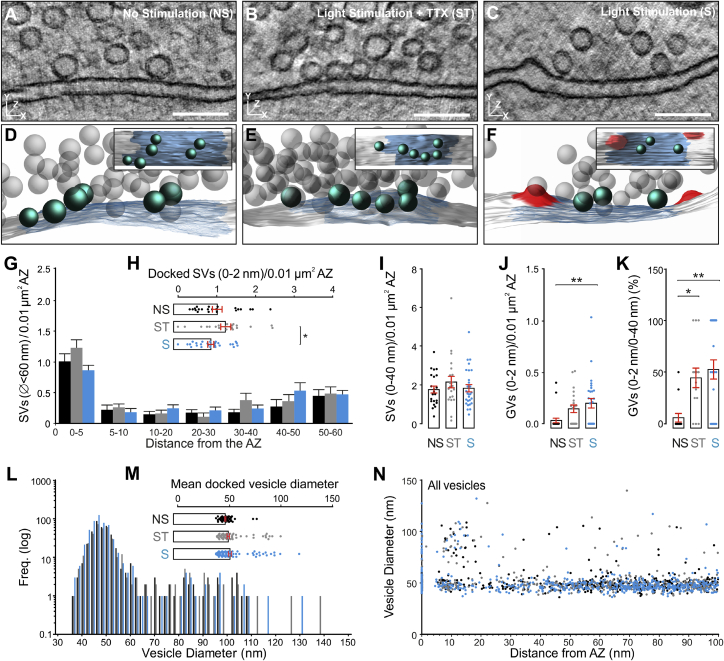
Figure 5.
3D Ultrastructural Analysis of Mossy-Fiber-CA3 Synapses in Dock10-Cre;Ai32 Slices after High-Frequency Stimulation (100 × 5 ms LPs at 20 Hz, 2 mM Ca2+)
(A–C) High-magnification electron tomographic subvolumes of active zones (AZs) for three conditions: NS, no stimulation (A); ST, stimulation in the presence of 1 μM TTX (B); S, stimulation (C).
(D–F) 3D models (docked vesicles, turquoise; undocked vesicles, gray; AZ, blue; putative endocytic pits, red) and orthogonal views (insets).
(G) Distribution of SVs (Ø < 60 nm) within 60 nm of the AZ normalized to AZ area.
(H) Spatial density of docked SVs (0- to 2-nm bin) normalized to AZ area.
(I) SV number within 0 to 40 nm of the AZ normalized to AZ area.
(J) Spatial density of docked giant vesicles (GVs; Ø >60 nm) normalized to AZ area.
(K) Percentage of docked GVs within 0 to 40 nm of the AZ.
(L) Vesicle diameter distribution for undocked vesicles within 100 nm of the AZ (2 nm bins). NS, 395 vesicles; ST, 365 vesicles; S, 516 vesicles.
(M) Mean diameter of docked vesicles. NS, 103 vesicles; ST, 115 vesicles; S, 108 vesicles.
(N) Spatial distribution of vesicles with respect to vesicle diameter.
Scale bars: (A–C) 100 nm. Error bars indicate mean ± SEM; ∗p < 0.05; ∗∗p < 0.01. NS, three cultures, three slices, n = 22 AZs; ST, three cultures, three slices, n = 21 AZs; S, two cultures, three slices, n = 28 AZs.