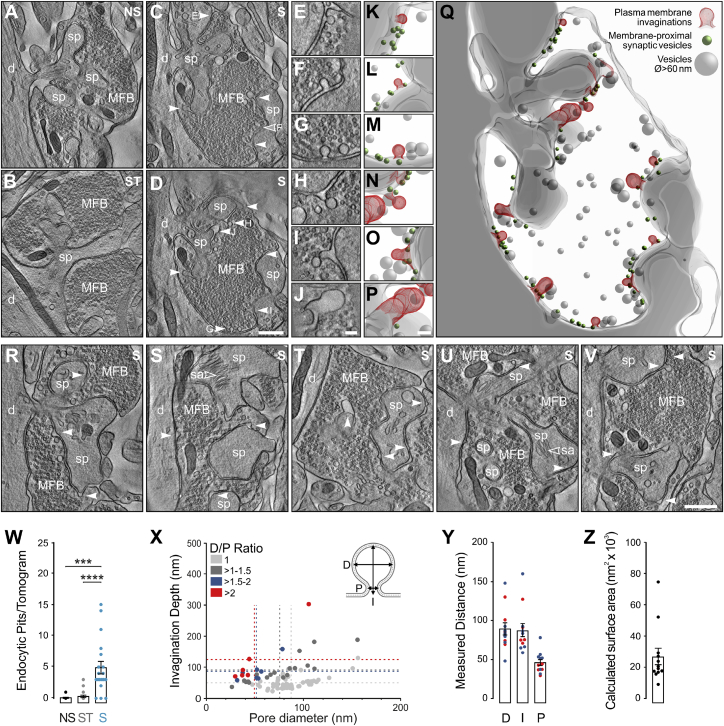
Figure 7.
3D Ultrastructural Analysis of Putative Endocytic Profiles in Dock10-Cre;Ai32 Slices after High-Frequency Stimulation (100 × 5 ms LPs at 20 Hz, 2 mM Ca2+)
(A–D) Low-magnification electron tomographic subvolumes. NS, no stimulation (A); ST, stimulation in the presence of 1-μM TTX (B); S, stimulation (C); tomographic slices from the same MFB (D). Mapping putative endocytic events that are within (white arrowheads) and outside the field of view (open white arrowhead).
(E–J) Manipulation of tomographic slice tilt angles revealed the shape and full extent of individual membrane invaginations from (C) and (D).
(K–Q) 3D models generated from individual events (E–J) and of the entire reconstructed tomographic volume (Q). Events (red) were frequently observed close to membrane-proximal vesicles (green). Vesicles of large diameter (Ø > 60 nm; gray spheres) can be found within the vicinity of endocytic events and may represent GVs or endocytosed vesicles.
(R–V) Further examples of endocytic events in stimulated MFBs.
(W) Number of putative endocytic profiles per tomogram.
(X) Relationship between pore diameter (P) and invagination depth (I) according to the ratio of maximum diameter (D) and pore diameter (D/p ratio = 1, light gray; D/p = > 1:1.5, dark gray; D/p = 1.5:2, blue; D/p = >2, red; respective mean values indicated by dashed lines).
(Y) Parameters (D, maximum diameter; I, invagination depth; P, pore diameter) pooled from endocytic events with D/P ratios of 1.5:2 (blue) and >2 (red) for pore diameters < 100 nm.
(Z) Calculated surface area of predicted endocytosed vesicles from profiles with D/P ratios >1.5.
Scale bars: (A–D and R–V), 1 μm; (E–P), 100 nm. Error bars indicate mean ± SEM; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. NS, one culture, one slice, n = 10 tomograms; ST, two cultures, two slices, n = 19 tomograms; S, two cultures, two slices, n = 19 tomograms.