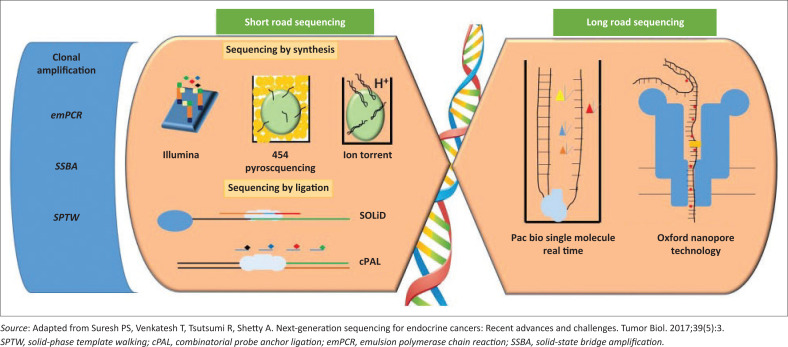
FIGURE 1.
Overview of short- and long-read sequencing technologies. Short-read sequencing methods are displayed on the left, long-read sequencing methods are shown on the right. Short-read sequencing methods are classified into sequencing by synthesis and sequencing by ligation. They all require clonal amplification; emulsion polymerase chain reaction for 454 pyrosequencing, SOLiD™ and Ion Torrent, SSBA is peculiar to Illumina, solid-phase template walking is used for certain SOLiD™ technologies. Illumina, fluorescent-labelled dNTP added to bridge amplified DNA template; 454 pyrosequencing, empolymerase chain reaction-generated microbead-bound DNA clone in picotitre well, DNA polymerase is added to the well, nucleotides are washed over in turn, deoxynucleoside triphosphate incorporation monitored via pyrophosphate release; Ion Torrent, similar to 454 pyrosequencing, deoxynucleoside triphosphate incorporation monitored via H+ ions release detected by a pH sensor that uses complementary metal-oxide-semiconductor technology. SOLiD™, microbead-bound DNA template flanked by adapters is hybridized to a growing complementary strand, under the action of DNA ligase. cPAL, another sequencing by ligation technique not described in this paper, employed by Complete Genomics, anchor sequence and probes hybridize to DNA template in a series of ligation reactions taking place on a nanoball. PacBio single-molecule real-time, sequencing takes place on zero-mode waveguide chip. DNA polymerase at the bottom of the well, fluorescent nucleotides being added to the strand. Oxford nanopore technology relies on changes in ion flow as nucleotides pass through the nanopore.