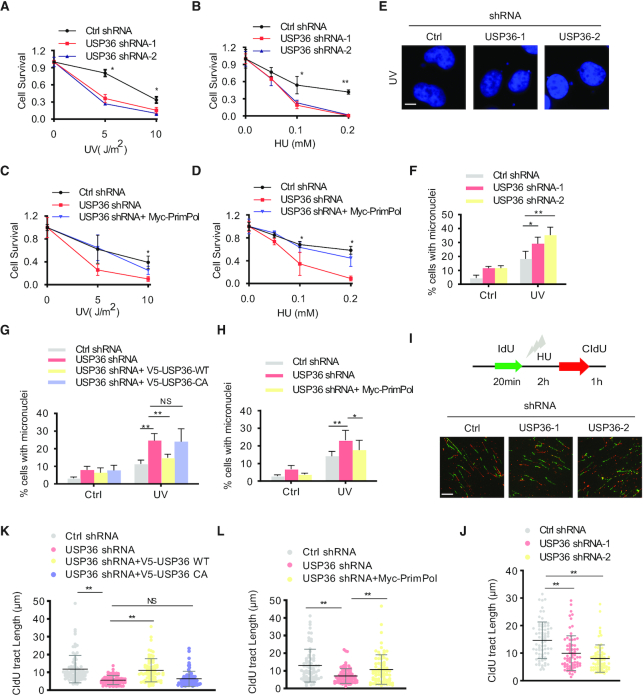
Figure 6.
USP36 participates in DNA replication stress through a PrimPol-dependent manner. (A, B) OVCAR8 cells from Supplementary Figure S7A were treated with indicated does of UV or HU. Cell survival was determined by colony formation assay. Error bars represent ± SD from three independent experiments. (C, D) OVCAR8 cells from Supplementary Figure S7B were treated with indicated does of UV or HU. Cell survival was determined by colony formation assay. Error bars represent ± SD from three independent experiments. (E) Representative images of micronuclei labeled with the DNA dye DAPI in OVCAR8 USP36-knockdown cells from Supplementary Figure S7A. Cells were left untreated or treated with UV (5 J/m2) for 48 h. Cells were fixed, permeabilized in 0.5% Triton X-100, stained with DAPI, then analyzed by Nikon eclipse 80i fluorescence microscope for micronuclei. The micronuclei assays were repeated for three times, and at least 200 cells were counted for each experiment. Scale bar in the bottom left is 10 μm. (F) Quantification of micronuclei in cells from E. (G) OVCAR8 USP36-knockdown cells reconstituted with V5-tagged USP36 (WT and C131A mutant) from Supplementary Figure S7C were used for measuring micronuclei. (H) OVCAR8 USP36-knockdown cells reconstituted with Myc-PrimPol from Supplementary Figure S7B were used for measuring micronuclei. (I) Representative images of DNA fibers in OVCAR8 USP36-knockdown cells from Supplementary Figure S7A. Cells were labeled with IdU for 20 minutes as a control to label the active replication, and followed by incubation with 4 mM HU for 2 h. Cells were then labeled with CIdU for 1h to evaluate the DNA replication upon genotoxic stress. DNA fibers were stretched on a microscope slide, stained with IdU and CIdU antibodies, imaged, and the lengths of fiber tracks measured. Scale bar in the bottom left is 10 μm. (J) Quantification of DNA fibers in cells from I. (K) OVCAR8 USP36-knockdown cells reconstituted with V5-tagged USP36 (WT and C131A mutant) from Supplementary Figure S7C were used to detect the restart efficiency of stalled replication forks. (L) OVCAR8 USP36-knockdown cells reconstituted with Myc-PrimPol from Supplementary Figure S7B were used to detect the restart efficiency of stalled replication forks. The graphs represent mean ± SD, two-tailed, Student's t-test. *P < 0.05; **P < 0.01; NS = not significant.