Table 2.
M.BceJIII and M.EcoGIX MTases, but not inactive variants suppress M13 replication and virion production in vivo.
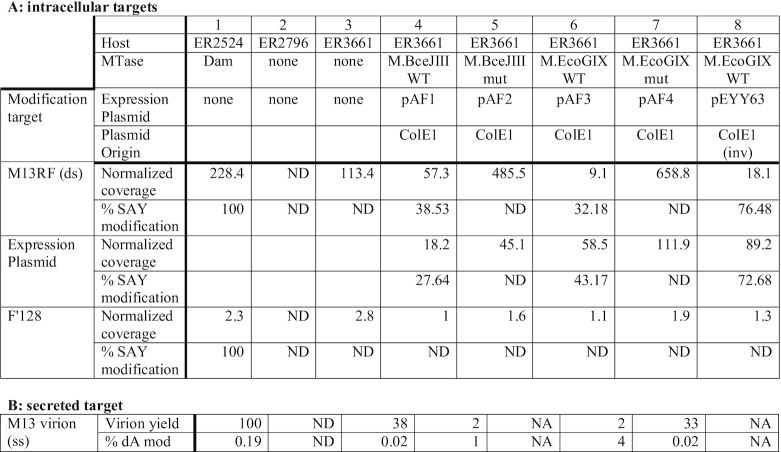
|
ND: not detected; NA: not applicable. Table 2A: Coverage: Number of PacBio reads mapped to reference sequences; % modification: SAY sites modified/SAY present x 100, except control host ER2524 column 1, Dam sites (GATC) modified/Dam sites present x 100. Expression plasmids were pAF1 (bceJIIIM WT); pAF2 (bceJIIIM mut); pAF3 (ecoGIXM WT); pAF4 (ecoGIXM mut); pEYY63 (ecoGIXM WT Ori inv). Table 2B: virion DNA was prepared by infection of ER2524 (Dam-modified host), ER2796 (non-modifying F- non-host), ER3661 (non-modifying host), and ER3661 transformed with pAF4 (ecoGIXM mut), pAF3 (ecoGIXM WT) or pAF1 (bceJIIIM WT); M13 virion yield was estimated from Qubit reads of preparations and expressed relative to the Dam+ control host ER2594 taken as 100%. Fraction of modified dA residues in the preparations was determined as by LC-MS described in Materials and Methods.
