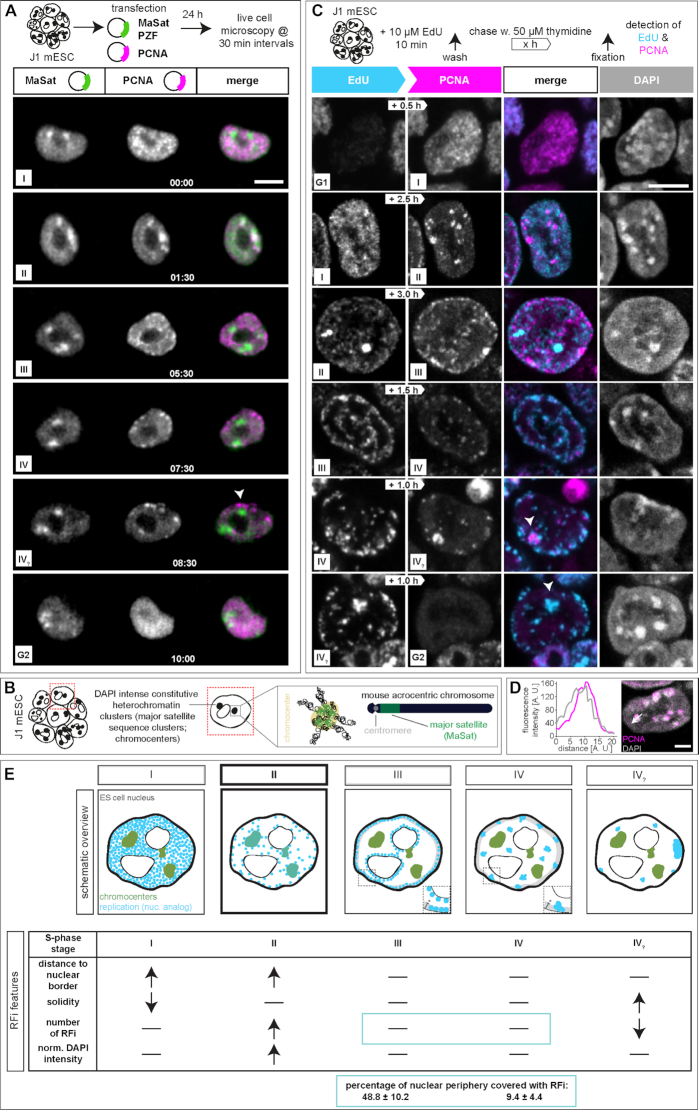
Figure 1.
DNA replication dynamics in mouse embryonic stem cells. (A) Experimental setup of a live cell experiment to determine the in vivo spatio-temporal progression of DNA replication in J1 mES cells. Cells were transfected with plasmids encoding mRFP-PCNA (magenta) and GFP-tagged polydactyl zinc finger protein (PZF) specifically binding to major satellite repeats (MaSat-GFP, green) fusion constructs to mark ongoing DNA replication and pericentromeric heterochromatin (chromocenters), respectively. Imaging was performed for 24 h with 30 min intervals. Representative spinning disk confocal images show the cell cycle progression of a representative mES cell and S-phase was further subdivided into five main replication patterns (I–IV?). The arrowhead in the IV? stage marks a prominent accumulation of replication signals observed at the end of S-phase. (B) Schematic representation of acrocentric mouse chromosome clustering in mES cell nuclei. At the chromosomal level, constitutive heterochromatin major satellite repeats (green) flank the centromere (grey) and in interphase nuclei, pericentromeric DNA from different chromosomes clusters to chromocenters. (C) Experimental setup of a pulse-chase experiment to determine the spatio-temporal progression of DNA replication in mouse J1 ES cells. Asynchronously growing mES cell cultures were pulse labeled with the nucleotide analog EdU, followed by various thymidine chase periods (white arrows) and fixation. EdU, i.e. nascent DNA during the first pulse labeling (cyan), and endogenous PCNA, i.e. ongoing replication at the time point of fixation (magenta), were (immuno)fluorescently detected and allowed the identification and the temporal order classification of five main replication patterns in mouse J1 ES cells. Representative spinning disk confocal images of G1 to S-phase, S-phase substage transitions (I–IV?) and S-phase to G2 progression are shown. The arrowheads in the IV? stage mark a prominent accumulation of replication signals observed at the end of S-phase. (D) Line profile analysis of PCNA fluorescence intensities within one chromocenter in a stage II cell. (E) Schematic summary of the five replication patterns observed in mES cells. Replication signals are shown in cyan, pericentromeric heterochromatin in green and replicating chromocenters (stage II) are marked in dark cyan. Table summarizing the significant RFi features of the five S-phase substages. Grey areas in stage III and IV S-phase schematic cells represent the nuclear periphery segmented to determine the percentage of the latter covered with RFi. The width of this area (*) corresponds to the diameter of stage III peripheral RFi (0.44 ± 0.13 μm). All experiments were done in at least three independent biological replicates. Detailed statistics are summarized in Supplementary Tables S7 and S11. Scale bars = 5 μm. Dotted lines represent cell contours.