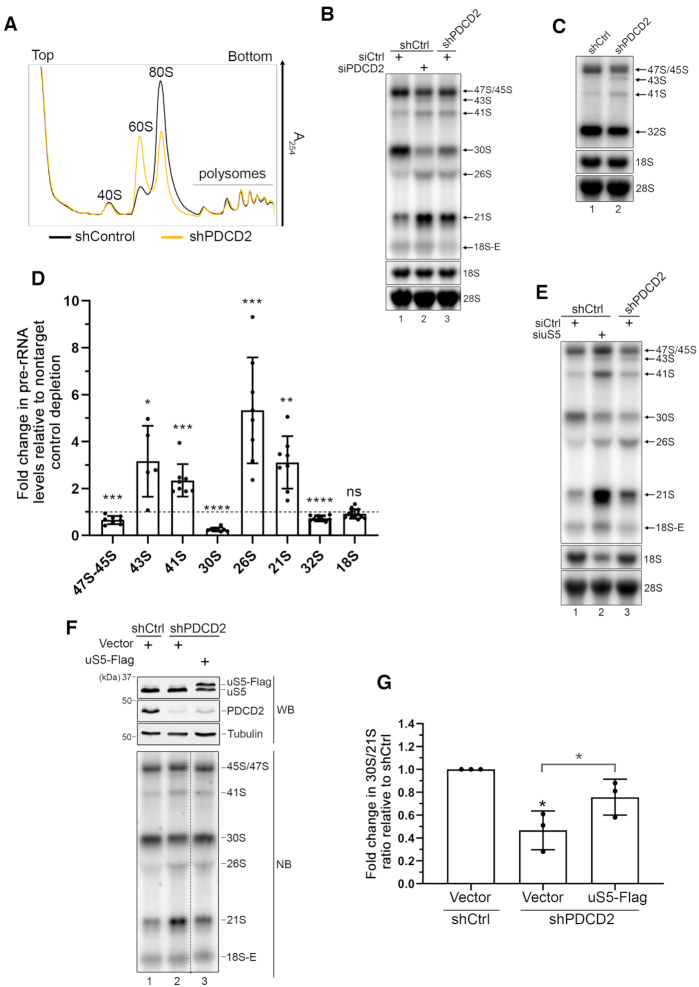
Figure 2.
PDCD2 contributes to 40S ribosomal subunit biogenesis. (A) Sucrose gradient analysis of total extracts prepared from HeLa cells that were previously induced to express a control nontarget (black profile) or a PDCD2-specific (yellow profile) shRNA for 96 h. The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), and polysomes are indicated. (B) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lanes 1–2) or a PDCD2-specific (lane 3) shRNA and that were transfected with nontarget (lanes 1 and 3) or PDCD2-specific (lane 2) siRNA. Pre-rRNAs were detected by using a probe complementary to sequences in the ITS1 region. Pre-rRNA species are indicated on the right. (C) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lanes 1) or a PDCD2-specific (lane 2) shRNA. Pre-rRNAs were detected by using a probe complementary to sequences in the ITS2 region. Pre-rRNA species are indicated on the right. (D) Levels of the indicated pre-rRNA were normalized to the mature 28S rRNA and are expressed relative to values for cells treated with a nontarget control depletion. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value > 0.05; *P-value <0.05; **P-value <0.01; ***P-value <0.001; ****P value <0.0001, as determined by Student's t test. (E) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lanes 1–2) or a PDCD2-specific (lane 3) shRNA and that were transfected with nontarget (lanes 1 and 3) or uS5-specific (lane 2) siRNA. Pre-rRNAs were detected by using a probe complementary to sequences in the 5′ ITS1 region. Pre-rRNA species are indicated on the right. (F) Western blot (WB, Top) and Northern blot (NB, Bottom) analysis from HeLa cells induced to express a nontarget control (lane 1) and PDCD2-specific (lanes 2–3) shRNA before transfection with a control plasmid (lanes 1–2) or a DNA construct expressing a Flag-tagged version of uS5 (lane 3). Pre-rRNAs were detected by using a probe complementary to sequences in the ITS1 region. Bottom panel: the data in lanes 1 to 3 were from the same blot, with the dashed line indicating some intervening lanes that were cropped out. (G) Quantification analysis of 30S/21S pre-rRNA ratios as determined by northern blot analysis (as F) from three independent experiments. The fold change in 30S/21S pre-rRNA ratio in PDCD2-depleted cells is presented relative to cells expressing a control nontarget shRNA (shCtrl). *P-value <0.05, as determined by Student's t test.