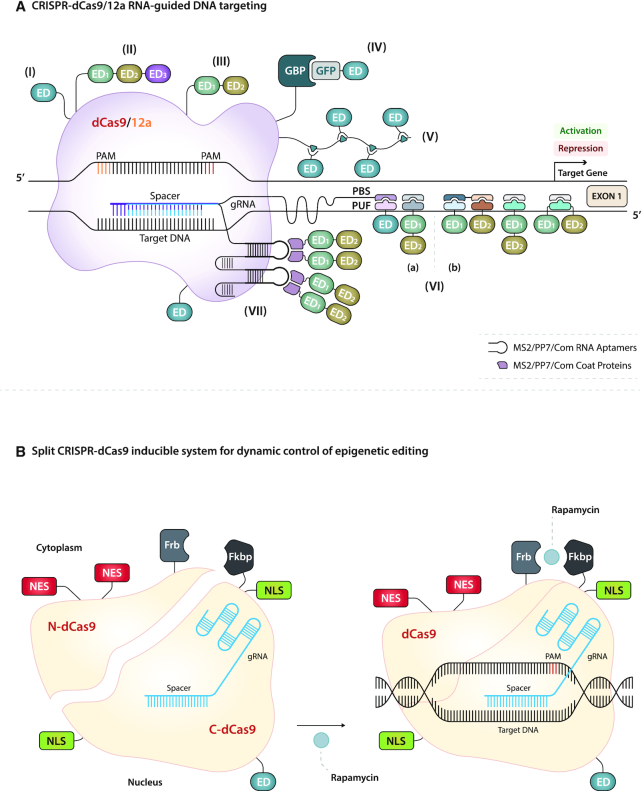
Figure 3.
CRISPR-dCas9 and 12a proteins for epigenome engineering. (A) dCas9 and 12a are guided to the DNA by a customizable guide RNA (gRNA) and CRISPR RNA (crRNA), respectively. The spacer is the interchangeable portion of the gRNA and crRNA that is complementary to the targeted DNA sequence, which is 20 nucleotides (blue) and 24 nucleotides (violet) in length for dCas9 and dCas12a, respectively. In order to recognize and bind the genomic sequence, dCas proteins also require a protospacer adjacent motif (PAM) immediately 3′ (red) and 5′ (orange) of the target DNA, for dCas9 and dCas12a, respectively. (I) A single ED directly fused to either the N- or the C-terminus of dCas. (II and III) A tripartite and a bipartite ED system directly linked to either the N- or the C-terminus of dCas. (IV) A modular recruitment system based on green fluorescent protein (GFP)-coupled ED via GFP-binding protein (GBP) fused to dCas proteins. (V) The SunTag system fused to dCas proteins for augmentation of epigenetic editing. (VI) The Casilio recruitment platform comprises an appended gRNA fused with one to five copies of Pumilio/FBF (PUF) binding sites (PBS) to recruit multiple distinct EDs, fused to a PUF domain. (a) Simultaneous gene activation and gene repression via EDs, independently recruited, by separate dCas9 proteins targeting different promoters within the same cell. (b) Enhanced gene activation via synergistic activities of distinct EDs recruited, in different combinations, via the same dCas9 protein. (VII) The Synergistic Activation Mediator (SAM) and gRNA 2.0 technology is based on a gRNA modified with MS2, PP7, or Com RNA aptamers from bacteriophages, which recruit EDs fused to aptamer coat proteins to enhance the epigenetic editing activity of dCas proteins already fused to EDs. (B) Schematic representation of the split dCas9 strategy for chemical induction of epigenetic editing. The two split fragments, N-dCas9 and C-dCas9 fused to ED, are joined to the rapamycin-binding domains Frb and Fkbp, respectively. Spatial sequestration inside the cell is maintained by an equal ratio of nuclear export sequences (NES) and nuclear localization sequences (NLS) separately fused to the two segments. The addition of rapamycin activates rapid and reversible dCas9 dimerization, thereby allowing dynamic control of transcriptional modulation.