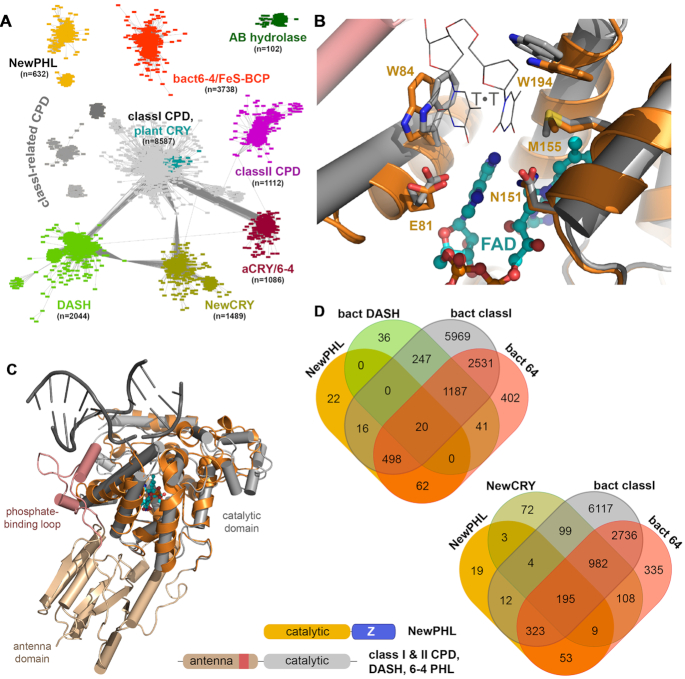
Figure 1.
Bioinformatics analysis of the whole photolyase-cryptochrome superfamily. (A) Sequence-similarity network of the PCSf including a combined set of sequences from PFAM families PF00875, PF04244 and INTERPRO family IPR005101. Each of the 9136 nodes represents sequences of a pairwise sequence of >50% (overall 19524 sequences), connecting edges represent BLAST E-value scores of <10−70. Two novel, exclusively bacterial PCF families have been identified by this SSN, NewCRY and NewPHL. Notably, class III CPD photolyases are an unresolved subset within the cluster for class I CPD photolyases and plant cryptochromes. A small cluster designated as AB hydrolases represents plant gene products, which harbour an N-terminal domain similar to the antenna domain of the PCSf and a C-terminal α/β-hydrolase domain. (B) Homology model of the NewPHL ortholog from Dinoroseobacter shibae (orange) that is superimposed to the template structure of the class I CPD photolyase of Synechococcus elongatus (PDB code: 1TEZ) in complex with substrate CPD-DNA (grey, sequence identity 30%). Its predicted active site structure of DsNewPHL (orange) harbours all residues required for CPD repair activity by SynCPDI (gray). Furthermore, the C4-carbonyl groups of the 5′- and 3′-thymines are predicted to form hydrogen bonds with the N6 amino group of the FADH− cofactor. (C) Overall view of the DsNewPHL catalytic domain in comparison to CPD-DNA bound state of SynCPDI. (D) Venn diagrams showing species (co)distribution between PCSf subfamilies with members of bacterial origin. A schematic representation of the domain architecture of the NewPHLs in comparison to canonical photolyases is shown in the lower part of the figure.