Figure 4.
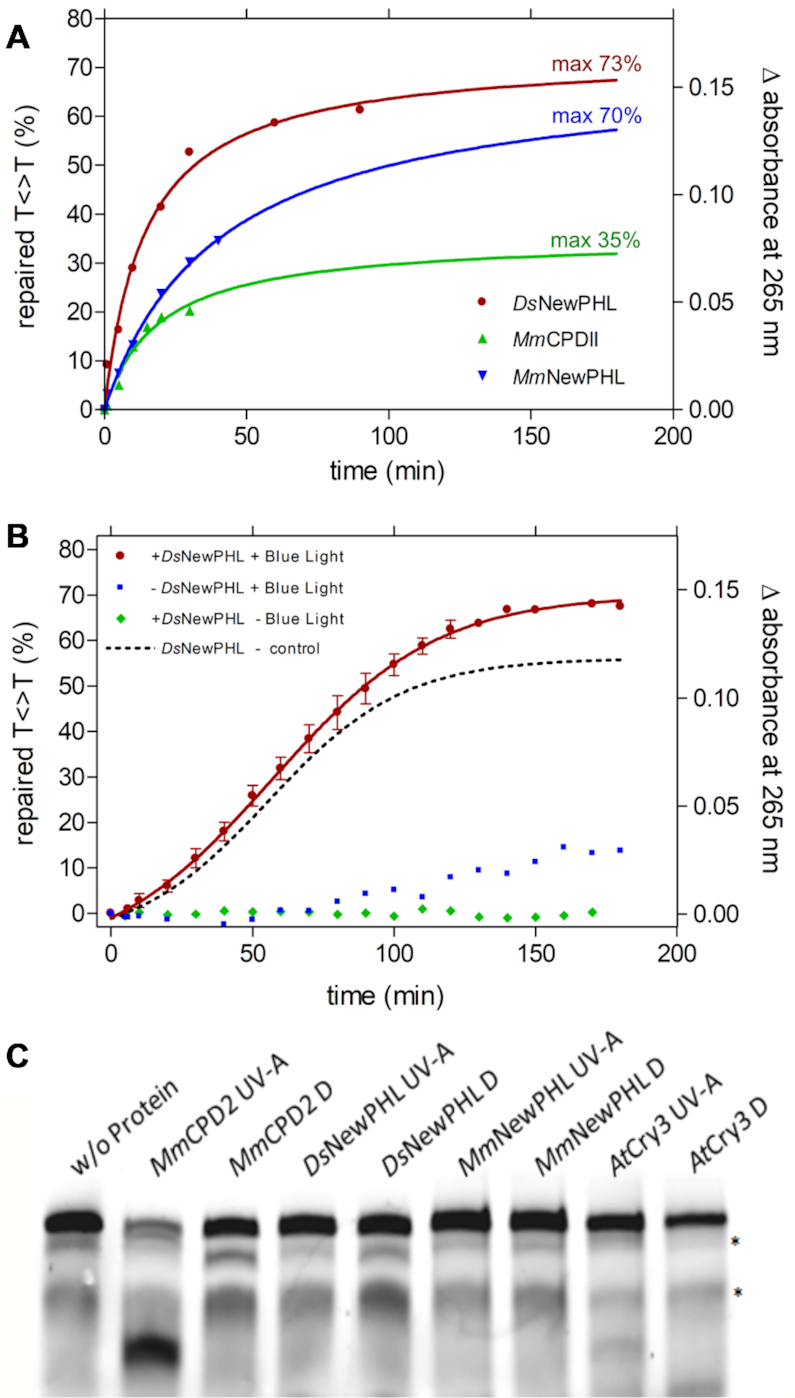
Repair activity and Restriction-site restoration assay of NewPHLs. Repair reactions contained 1 μm (A) or 100 nm (B) previously reduced protein, 5 μm UV-irradiated oligo(dT)18 and 0.175 mm DTT in a volume of 200 μl. Repair of CPD lesions was triggered with blue light (455 nm, 224 μmol·m−1·s−1) at 10°C and followed by UV-Vis spectroscopy. The difference in absorbance at 265 nm was plotted. Reaction-sample without protein and with no illumination served as a control (B). In (A) data points were extrapolated and the maximum repair was determined using an exponential fit. Interestingly, a slight absorption increase under illumination but without added enzyme can be observed after ∼80 min (B) that is caused by onset of DTT oxidation. For comparison the dotted curve shows light-dependent CPD-lesion repair corrected by this background reaction. (C) Restriction-site restoration assay with a single thymine cyclobutane dimer localized in the center of a well formed DNA duplex. The respective DNA probe (50 nm) was incubated with the indicated purified enzymes (each in 500 nm concentration) in the repair reactions in the presence of external reductant DTT (10 mm). Reactions were kept either in the dark (Dark) or under UV-A irradiation (+UV-A, 385 ± 5 nm, 25 μmol·m−2·s−1) for 60 min, and afterwards treated with VspI, denatured and separated on 4–15% polyacrylamide gels containing 7 m urea. The resulting bands were visualized using the Odyssey LiCOR Infrared Imaging system. The upper band shows the uncut substrate, the prominent lower band in the MmCPDII lane represents the repaired substrate cleaved by VspI;* unspecific band.
