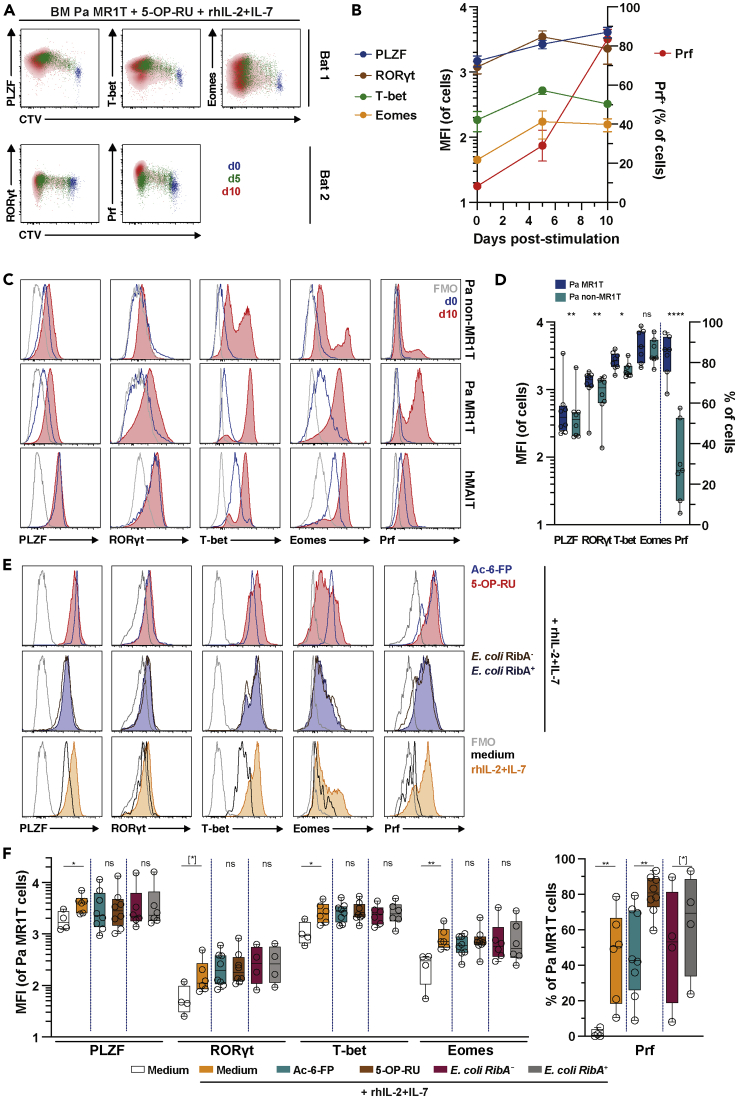
Figure 3.
Pa MR1T Cells Upregulate Effector Transcription Factors Following Proliferation.
(A and B) (A) Flow cytometry plots and (B) expression of the transcription factors PLZF, T-bet, and Eomes as well as RORγt and the cytolytic protein Prf from separate, independent experiments, respectively, along with CTV dilution by Pa MR1T cells on day 0, 5, and 10 following stimulation with 5-OP-RU supplemented with rhIL-2+IL-7 (n = 4-9).
(C and D) (C) Histograms of the transcription factors PLZF, RORγt, T-bet, and Eomes, and the cytolytic protein Prf at resting and following culture with 5-OP-RU supplemented with rhIL-2+IL-7 and (D) expression of the same effector transcription factors and cytolytic protein by Pa MR1T cells and Pa non-MR1T cells on day 10 of culture (n = 7-8).
(E and F) (E) Histograms and (F) levels (MFI) of the transcription factors PLZF, RORγt, T-bet, and Eomes, and the cytolytic protein Prf at day 7–10 of culture in medium alone, medium supplemented with rhIL-2+IL-7, or with various MR1 ligands, riboflavin-producing or -deficient E. coli strains in the presence of rhIL-2+IL-7 (n = 47-8). Data presented as a line graph with error bars represent the mean and standard error. Box and whisker plot shows all data points, median, and the interquartile range. Statistical significance was determined using the paired t test (RORγt, Eomes, Prf) (D), or Wilcoxon's signed-rank test (PLZF and T-bet) (D), between Ac-6-FP and 5-OP-RU (F), and between E. coli RibA+ and RibA− strains (F), or Mann-Whitney's test between medium and rhIL-2+IL-7 (F). ∗∗∗∗p < 0.0001, ∗∗p < 0.01,∗p < 0.05, [∗] p < 0.1. FMO, fluorescence minus one; ns, not significant.