Summary
Due to the lack of effective early diagnostic measures and treatment methods, bladder cancer has become a malignant tumor that seriously threatens people's lives and health. Here, we reported that LINC00162, a super-enhancer long noncoding RNA, was highly expressed in bladder cancer cells and tissues. And LINC00162 was negatively correlated with neighboring PTTG1IP expression. Knocking down LINC00162 expression can inhibit the proliferative activity of bladder cancer cells and the growth of transplanted tumors in vivo, while knocking down the expression of PTTG1IP could restore the proliferative activity of bladder cancer cells. In addition, both LINC00162 and PTTG1IP were found to be able to bind to THRAP3, a transcription-related protein. And THRAP3 can regulate PTTG1IP expression. Finally, we demonstrated a mechanism that LINC00162 could regulate PTTG1IP expression through binding THRAP3. This study provided a potential target molecule for clinical treatment of bladder cancer.
Subject Areas: Molecular Biology, Cancer
Graphical Abstract
Highlights
-
•
Expression of LINC00162 is increased in bladder cancer
-
•
LINC00162 promotes bladder cancer progress in vitro and in vivo
-
•
LINC00162 regulates neighboring PTTG1IP expression to promote bladder cancer
-
•
LINC00162 inhibits PTTG1IP expression by interacting THRAP3
Molecular Biology; Cancer
Introduction
Bladder cancer is a common malignant tumor of the urinary system. In 2019, there were an estimated 80,470 cases of bladder cancer in the United States, and there will be 17,670 deaths (Siegel et al., 2019). Bladder cancer is one of the ten most common malignant tumors. It has the highest incidence in the urinary system. Increasing morbidity and mortality each year has seriously threatened people's lives and health. However, with the rapid development of molecular diagnostic technology, a large number of tumor markers have been discovered, and the development of molecular targeted drugs has become an effective measure for tumor treatment. Therefore, the exploration of novel target molecules for bladder cancer has important clinical significance for the treatment of bladder cancer.
In recent years, noncoding RNA has gradually become a hot topic of research in medical research, and many studies have proved that noncoding RNA could participate in regulating the pathological processes of various cancers (Huang et al., 2016; Liang et al., 2018; Yuan et al., 2014; Zhang et al., 2019). There are 3 billion base pairs in the genome but only about 30% of transcripts derived from the genome can encode proteins, the remaining about 70% are noncoding RNA (Atianand and Fitzgerald, 2014; Parasramka et al., 2016). Although the number of noncoding RNA is much greater than that of coding RNA. However, the research on the biological function of noncoding RNA is still limited. Long noncoding RNA (lncRNA) is a type of long noncoding RNA longer than 200nt (Zhang et al., 2017). Although lncRNA itself does not have the function of encoding proteins, it could participate in regulating various cell physiological activities such as cell differentiation, proliferation, apoptosis, and metastasis (Xie et al., 2017). At present, studies have shown that lncRNA is abnormally expressed in many tumors and has become a potential target molecule for many cancers (Kong et al., 2015; Tan et al., 2018; Xiao et al., 2018; Xie et al., 2018). Moreover, lncRNA could also regulate the occurrence and development of malignant tumors through various pathways. For example, lncRNA NORAD competitively adsorbs hsa-miR-125a-3p through the sponge function and then regulates the expression of RhoA, thereby promoting the development of pancreatic cancer (Li et al., 2017). lncRNA TROJAN accelerates the degradation of ZMYND8 through the ubiquitination pathway and subsequently promotes the progression of breast cancer (Jin et al., 2019). lncRNA TUG1 could bind to the enhancer of EZH2, regulating the expression of LIMK2b, and then promote the cell growth and chemical resistance of small cell lung cancer (Niu et al., 2017). At present, various lncRNAs have been reported in literatures, such as lncRNA HIF1A-AS2, lncRNA host gene 16, and lncRNA SNHG16, which play a key role in the biological process of bladder cancer (Chen et al., 2019; Feng et al., 2018; Peng and Li, 2019).
We found that the expression of LINC00162 was most significantly upregulated in the super-enhancer lncRNA microarray expression profile. Compared with normal bladder epithelial cells and tissues, LINC00162 also had significant differences in bladder cancer cells and tissues. This study mainly explored the function of LINC00162 in the development of bladder cancer. We found that knocking down the expression of LINC00162 could effectively inhibit the proliferative viability of bladder cancer cells, the ability to form clones in soft agar, and the rate of tumor growth in nude mice. Therefore, LINC00162 plays a tumor-promoting function in bladder cancer. By studying the mechanism of LINC00162 regulating the proliferation of bladder cancer cells, we found that LINC00162 could inhibit the neighboring PTTG1IP expression through binding to THRAP3, a transcriptional regulatory protein, and finally promote the proliferation of bladder cancer cells. This discovery not only allowed us to understand the function and mechanism of LINC00162 in bladder cancer but also provided a potential target molecule for the treatment of bladder cancer.
Results
Detection of LINC00162 Expression in Bladder Cancer Cells and Tissues
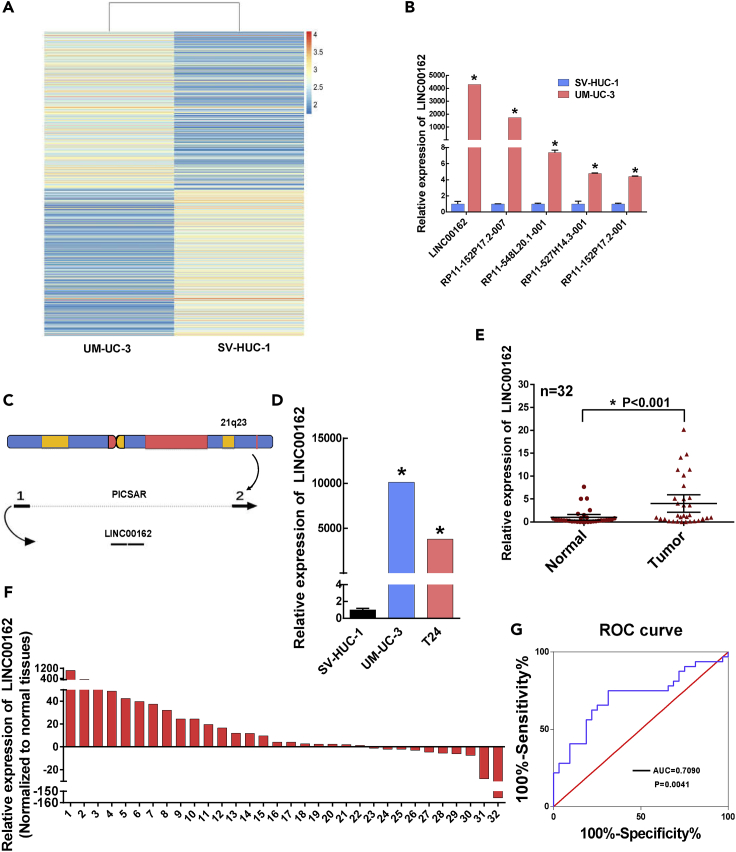
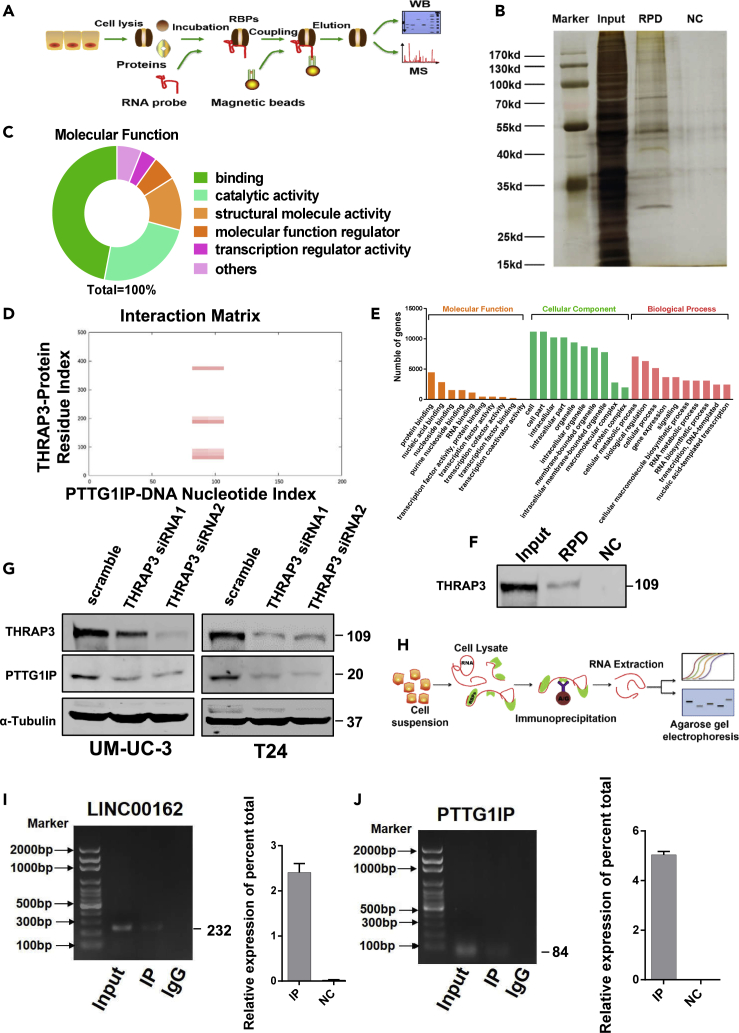
We used the immortalized normal bladder epithelial cell line SV-HUC-1 and bladder cancer cell UM-UC-3 as cell samples for super-enhancer lncRNA microarray analysis. We used fold changes in microarray analysis results to show differences in lncRNA expression (Figure 1A). Specific primers were designed to verify differentially expressed SE lncRNA between SV-HUC-1 and UM-UC-3 cells. And we verified five upregulated SE lncRNAs in UM-UC-3 cells by quantitative polymerase chain reaction (q-PCR) test, of which LINC00162 has the highest fold change (Figure 1B). So, LINC00162 was selected out to further explore its biological function and mechanism in vivo and in vitro. LINC00162 is composed of two exons of P38 inhibited cutaneous squamous cell carcinoma associated lincRNA (PICSAR) (Figure 1C). Then, we detected the expression of LINC00162 in SV-HUC-1, UM-UC-3, and T24 cells. Compared with SV-HUC-1 cells, the expression of LINC00162 in UM-UC-3 and T24 was significantly upregulated (Figure 1D). We also tested the expression of LINC00162 in 32 pairs of bladder cancer tissues and adjacent tissues. Compared with adjacent tissues, LINC00162 was upregulated in bladder cancer tissues (Figures 1E and. 1F). Subsequently, we used receiver operating characteristic curve (ROC) curve analysis and evaluation. The results showed that the area under the curve was 0.709 (95% confidence intervals (CI): 0.579–0.839), indicating that LINC00162 has a potential diagnostic value (Figure 1G).
Figure 1.
Detection of LINC00162 Expression in Bladder Cancer Cells and Tissues
(A) Heatmap analysis results of super-enhancer lncRNA microarray in the UM-UC-3 cell and the SV-HUC-1 cell.
(B) The expression of five upregulated lncRNAs in microarray analysis was detected in SV-HUC-1 and UM-UC-3 cells by q-PCR, ∗p < 0.05.
(C) Genomic structure of LINC00162. LINC00162 is composed of two exons of PICSAR, and PICSAR is located in the chromosome 21q22 region.
(D) Relative expression of LINC00162 in bladder normal epithelial cells and two bladder cancer cell lines. Data are expressed as mean ± standard deviation, n = 3, unpaired t test, ∗p < 0.05.
(E) The scatterplot shows the relative expression of LINC00162 (standardized according to GAPDH expression level) in 32 pairs of bladder cancer and adjacent tissues, n = 32, paired t test, ∗p < 0.05.
(F) Relative expression of LINC00162 in bladder cancer tissues of 32 patients relative to adjacent tissues.
(G) Receiver operating characteristic curve analysis to evaluate the diagnostic value of LINC00162. The area under the curve (AUC) was 0.709 (95% CI: 0.579–0.839).
LINC00162 Promotes Proliferation of Bladder Cancer Cells
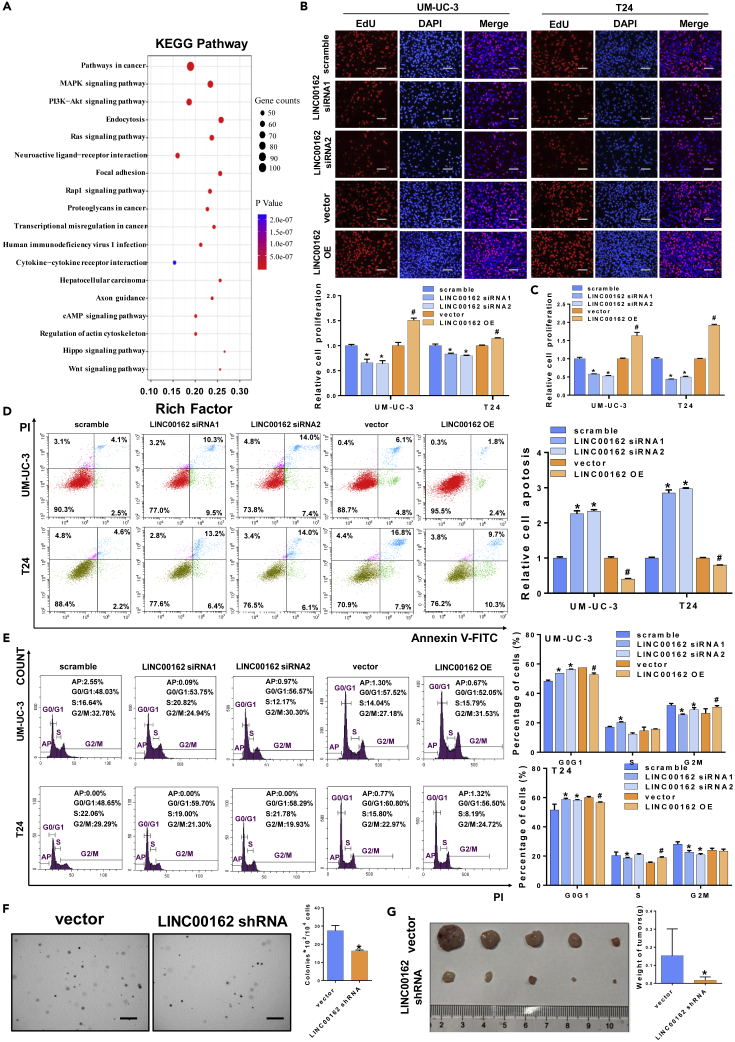
In order to initially predict the potential functions of LINC00162, we analyzed the RNAs regulated by LINC00162 and its pathways with bioinformatics methods. In brief, we first predicted the miRNA that LINC00162 might bind to through the RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/) and then predicted the mRNA that these miRNAs may bind to through the TargetScan (http://www.targetscan.org/vert_71/). Then, we conducted pathway enrichment analysis on these mRNAs and found that LINC00162 was involved in a variety of tumor-related pathways and was also enriched in pathways such as protein binding and biological regulation (Figures 2A and S1A–S1C). To further investigate whether LINC00162 is involved in the development of bladder cancer, we designed the siRNA of LINC00162 and constructed its overexpression plasmid and performed efficiency verification in UM-UC-3 and T24 cells (Figure S1D). Subsequently, to study the effect of LINC00162 on cell physiology, we silenced or overexpressed LINC00162 and found that various physiological behaviors of cells changed significantly, such as cell proliferation (Figures 2B and 2C), cell apoptosis (Figure 2D), and cell cycle (Figure 2E). Compared with the control group, UM-UC-3 and T24 cells transfected with the siRNA group (LINC00162 siRNA1 or LINC00162 siRNA2) significantly reduced cell viability, inhibited cell proliferation, promoted apoptosis, and increased cells in the G0/G1 block. Compared with the vector group, UM-UC-3 and T24 cells transfected with the LINC00162 overexpression plasmid significantly increased cell viability, promoted cell proliferation, inhibited apoptosis, and reduced G0/G1 phase block (Figures 2B–2E). In order to investigate the effect of LINC00162 in vivo, we constructed a stable knockdown cell line of LINC00162 shRNA using UM-UC-3 cells and tested the silencing efficiency of stable cells by q-PCR (Figures S2A–S2C). The number of monoclonal cells in the LINC00162 shRNA group was significantly reduced compared with the control group in the soft agar clone formation experiment (Figure 2F). Next, we studied the effect of LINC00162 on tumor-bearing in nude mice. LINC00162 shRNA stable cell lines and vector stable cell lines were subcutaneously injected into nude mice, and the tumors were removed after 21 days for observation and weighing (Figure 2G). We found that tumors formed by LINC00162 shRNA stable cell lines were significantly decreased compared to the control group.
Figure 2.
LINC00162 can Promote the Proliferation of Bladder Cancer Cells
(A) LINC00162 KEGG pathway analysis. The vertical axis represents the pathway. The horizontal axis represents the enrichment factor (ratio of the number of annotated genes in the corresponding pathway rich in differentially expressed genes). The size of the dots indicates the number of differentially expressed genes in the corresponding pathway (number of genes). The color of the dots indicates the range of p values.
(B) The 5-ethynyl-2’-deoxyuridine (EdU) assay was used to detect the role of LINC00162 in bladder cancer cell proliferation, and the scale bar represents 300 μm∗p < 0.05.
(C) The role of LINC00162 in the activity of bladder cancer cells was detected by the ATP method, ∗p < 0.05.
(D) Flow cytometry was used to detect the role of LINC00162 in bladder cancer cell apoptosis, ∗p < 0.05.
(E) The function of LINC00162 in bladder cancer cell cycle was tested by flow cytometry. Experimental cells were stained with propidium iodide (PI). Cell counts represent the number of cells at different cell cycle stages, ∗p < 0.05.
(F) The results of the soft agar experiment (UM-UC-3-vector and UM-UC-3-LINC00162-shRNA), and the scale bar represents 200 μm, ∗p < 0.05.
(G) Subcutaneous tumor-bearing results in nude mice at 27 days, ∗p < 0.05.
LINC00162 Affects Neighboring Gene Expression
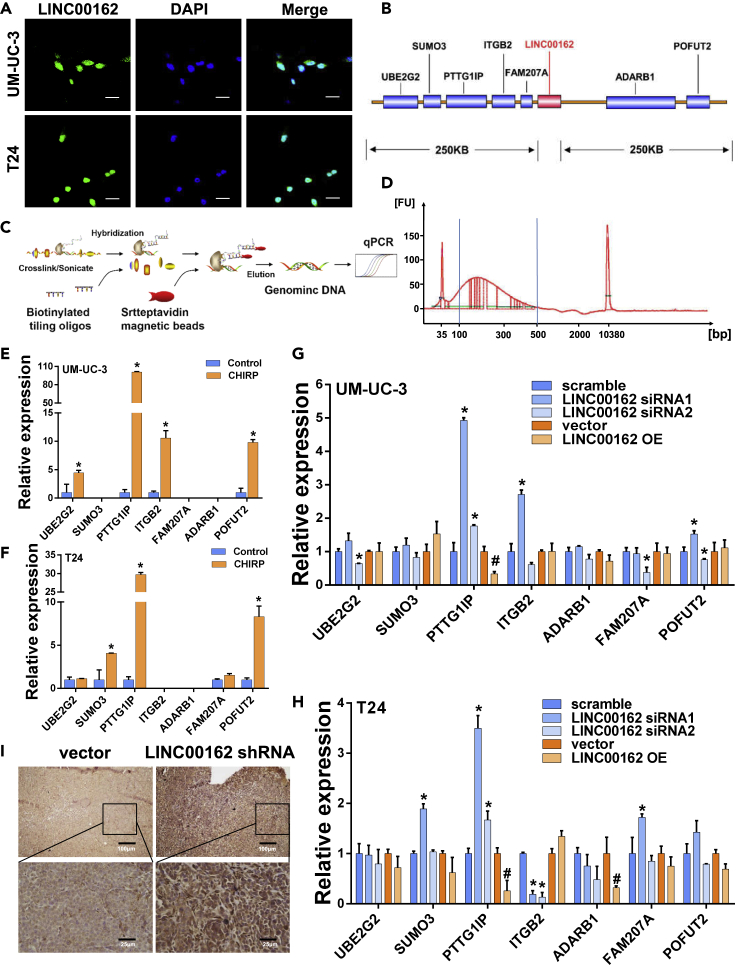
To illustrate the distribution of LINC00162 at the subcellular level, we performed fluorescence in situ hybridization experiments (FISH) in UM-UC-3 and T24 cells. As shown in Figure 3A, LINC00162 is mainly expressed in the nucleus. By comparing the chromosomal position of LINC00162, it was found that the sequence encoding LINC00162 overlaps with the segment encoding the super-enhancer. To investigate whether LINC00162 can regulate neighboring genes expression, we selected neighboring genes in the range of 250KB upstream and downstream of LINC00162, respectively UBE2G2, SUMO3, PTTG1IP, ITGB2, FAM207, ADARB1, POFUT2 (Figure 3B). Subsequently, we designed an RNA-specific probe for LINC00162, and according to the requirements of the chromatin isolation by RNA purification (CHIRP) experiment (Figure 3C), it ultrasonically shattered the whole genome DNA in UM-UC-3 and T24 cells. So, the content of the fragment was more than 70% at 100bp-500bp (Figure 3D). Next, the DNA fragment bound to LINC00162 was pulled down through the CHIRP experiment. q-PCR experiments showed that compared with other neighboring genes, PTTG1IP was bounded and pulled down by LINC00162 probes (Figures 3E and 3F). To determine whether there was an expression correlation between LINC00162 and neighboring genes, we used LINC00162 siRNA1 and siRNA2 to silence the expression of LINC00162 RNA. The transient LINC00162 OE plasmid was used to overexpress LINC00162 expression. As shown in Figures 3G and 3H, the correlation between PTTG1IP and LINC00162 expression was highest in UM-UC-3 and T24 cells, and it showed a significant negative correlation, suggesting that LINC00162 is an upstream gene of PTTG1IP and regulates the expression of PTTG1IP. In order to further determine the regulatory role of LINC00162 on PTTG1IP in vivo, we fixed, embedded, and sliced the tumors formed in nude mice subcutaneous tumor experiments and performed immunohistochemical experiments to detect the expression of PTTG1IP. As shown in Figure 3I, the expression of PTTG1IP protein was increased in the LINC00162 knockdown tissue compared with the control group, suggesting that LINC00162 can inhibit the expression of PTTG1IP in vivo.
Figure 3.
LINC00162 Affects the Expression of Neighboring Genes
(A) FISH assay detected the subcellular localization of LINC00162. Images were acquired under a confocal fluorescence microscope. Green fluorescence represents 6-FAM labeled circRNA probes, and blue fluorescence represents DAPI-stained nuclei, and the scale bar represents 100 μm.
(B) The relative position of LINC00162 and adjacent genes (selected the adjacent 250kb gene above and below LINC00162).
(C) Schematic diagram of CHIRP experiment.
(D) DNA sample test results by Agilent 2100 for CHIRP experiments (100bp-500bp sample content> 70%).
(E and F) The results of CHIRP experiment samples were detected in UM-UC-3 cells and T24 cells by q-PCR, ∗p < 0.05.
(G and H) Effect of silenced and overexpressed LINC00162 on neighboring gene mRNA expression in UM-UC-3 cells and T24 cells, ∗p < 0.05.
(I) Differential expression of PTTG1IP proteins in tumors of nude mice was detected by immunohistochemistry.
LINC00162 Promotes Proliferation of Bladder Cancer Cells by Regulating PTTG1IP
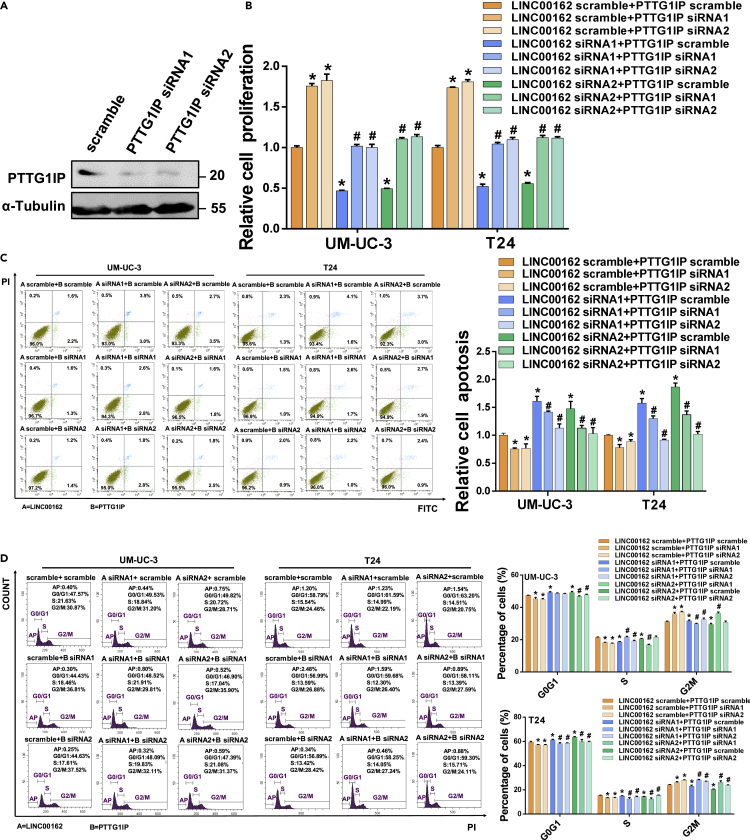
To investigate whether PTTG1IP is involved in the LINC00162 mechanism that promotes the proliferation of bladder cancer cells, we first designed and synthesized siRNA for PTTG1IP mRNA and verified the knockdown efficiency of siRNA on PTTG1IP proteins level (Figure 4A). Next, we determined the effects of LINC00162 and PTTG1IP on cell proliferation (Figure 4B), apoptosis (Figure 4C), and cycle (Figure 4D) by flow cytometry and measuring total adenosine triphosphate (ATP) content. Silencing the expression of LINC00162 alone could reduce the proliferation of bladder cancer cells, increase the apoptosis of bladder cancer cells, and block the G0/G1 phase. But silencing PTTG1IP expression alone could promote the proliferation of bladder cancer cells, reduce the apoptosis of bladder cancer cells, and promote G0/G1 phase. Compared with the control group, silencing the expression of LINC00162 combined with silencing the expression of PTTG1IP showed no statistically significant changes in bladder cancer cell proliferation, apoptosis, and cycle. These results showed that PTTG1IP could inhibit the proliferation of bladder cancer, promote cell apoptosis, and block G0/G1 phase and that LINC00162 could inhibit the regulation of PTTG1IP.
Figure 4.
LINC00162 Promotes the Proliferation of Bladder Cancer Cells by Regulating PTTG1IP
(A) Western blot verified the efficiency of PTTG1IP silencing.
(B) Effects of LINC00162 and PTTG1IP on the viability of UM-UC-3 and T24 cells detected by the ATP method, ∗p < 0.05.
(C) Effect of LINC00162 and PTTG1IP on apoptosis of UM-UC-3 and T24 cells. Annexin Fluorescein isothiocyanate isomer (FITC) and propidium iodide (PI) are used to indicate apoptosis, ∗p < 0.05.
(D) The effects of LINC00162 and PTTG1IP on the cell cycle of UM-UC-3 and T24. Experimental cells were stained with propidium iodide (PI). Count the number of cells at different cell cycle stages, ∗p < 0.05.
LINC00162 Regulates PTTG1IP by Binding to THRAP3
In order to investigate how LINC00162 regulates the expression of PTTG1IP, we used RNA pull-down experiments and mass spectrometry to investigate whether LINC00162 regulates PTTG1IP by recruiting transcription-related proteins. To this end, we designed an RNA pull-down specific probe for LINC00162 and performed an RNA pull-down experiment in the UM-UC-3 cell line (Figure 5A). Silver staining results showed that there were a variety of proteins that could bind to the LINC00162 molecule, the molecular weight distribution of the protein was relatively wide, and the protein enrichment was highest at 40kd-55kd and 100kd (Figure 5B). After protein purification, many related proteins were obtained by mass spectrometry analysis, and most of the proteins had binding functions (Figures 5C and S3A–S3C). We screened multiple transcription-related proteins and found that THRAP3 had the highest binding index with PTTG1IP DNA nucleotide (Figure 5D). We analyzed the molecular function of THRAP3 through gene ontology (GO) annotations and found that THRAP3 could participate in multiple transcriptional regulatory pathways (Figure 5E). To ensure the protein identity of THRAP3, we derived the protein map of THRAP3 using mass spectrometry (Figure S3D) and tested the content of THRAP3 protein in the input group, the RNA pull-down (RPD) group, and the negative control (NC) group by Western blot experiments. As shown in Figure 5F, the THRAP3 protein could indeed be pulled down by the specific molecular probe of LINC00162, and it proved that THRAP3 could bind to LINC00162 again. To investigate the correlation between THRAP3 and PTTG1IP, we performed THRAP3 expression silencing in two bladder cancer cell lines (UM-UC-3 and T24) and collected the proteins from the two cell lines for Western blot detection. As shown in Figure 5G, after the THRAP3 protein expression was downregulated, the PTTG1IP protein expression also decreased accordingly, suggesting that THRAP3 could positively regulate PTTG1IP expression. In order to further prove the binding relationship of THRAP3 with LINC00162 and PTTG1IP, we used the THRAP3 antibody to pull the bound molecule through RNA Immunoprecipitation (RIP) experiments (Figure 5H) and then performed PCR to detect candidate molecules. The contents of LINC00162 and PTTG1IP DNA in the input group, Immunoprecipitation (IP) group, and IgG group were showed by agarose gel electrophoresis (Figures 5I and 5J). The results indicated that the contents of LINC00162 and PTTG1IP DNA in the IP group were significantly higher than those in the IgG group, showing that THRAP3 has a binding effect with LINC00162 and PTTG1IP.
Figure 5.
LINC00162 Regulates PTTG1IP by Binding to THRAP3
(A) Schematic of RNA pull-down experiment.
(B) Silver-stained analysis of RNA pull-down experimental samples (RNA-binding proteins).
(C) The RNA-binding proteins detected by mass spectrometry were analyzed by Gene Ontology.
(D) Prediction of THRAP3 and PTTG1IP mRNA binding. Interaction tendencies and discrimination in the diagrams represent the possibility and ability of interaction, respectively.
(E) Gene Ontology annotations of THRAP3.
(F) Western blot was used to detect the protein content of THRAP3 in the input group, RPD group, and NC group in the pull-down experimental samples.
(G) The effect of changes in THRAP3 protein on the expression of PTTG1IP protein. THRAP3 siRNA was transfected into UM-UC-3 cells and T24 cells, respectively. Cell protein was collected 48 hr later, and the protein levels of THRAP3 and PTTG1IP were detected by Western blot experiments.
(H) Schematic of RIP experiment.
(I and J) Agarose gel electrophoresis was used to detect the content of LINC00162 or PTTG1IP mRNA in the input group, IP group, and IgG group in RIP experiments.
Discussion
Bladder cancer is a malignant tumor with the highest incidence in the urogenital system (Jeronimo and Henrique, 2014; Robertson et al., 2017). Because early symptoms are not obvious, most patients develop bladder cancer that reaches the middle or late stage when diagnosed, and this has greatly hindered clinical treatment (Antoni et al., 2017; Maia et al., 2018; Morales et al., 2016). Some progress has been made in bladder cancer research, and a large variety of tumor molecular targets have been found at the molecular level, such as genes and proteins (P53, Ki-67), providing important references for early diagnosis and treatment of bladder cancer (Bazrafshani et al., 2016; Noel et al., 2015; Onal et al., 2015; Puntoni et al., 2016). However, these molecular tumor targets that are used currently still fail to meet the ideal requirements in terms of sensitivity or specificity. Therefore, more effective tumor molecular targets are urgently needed for clinical diagnosis and treatment support, and research on new target molecules in bladder cancer is extremely important. LncRNAs are an important constituent member of the endogenous RNAs regulatory network (Zhang et al., 2018a, 2018b). In recent years, increasing evidence has suggested that lncRNAs can participate in the regulation of various levels of chromosome modification, RNA transcription, and protein translation in physiological cell processes (Hon et al., 2017; Park et al., 2018; Podbevsek et al., 2018). In addition, lncRNAs can also have important effects on various stages of cell physiology (Misawa et al., 2016; Roisman et al., 2019; Xu et al., 2019). At the present time, a large amount of literature has shown that lncRNAs can regulate the development of bladder cancer in various ways. For example, lncRNA MAGI2-AS3 adsorbs miRNA through the sponge function and then regulates the protein expression of CCDC19, thereby inhibiting the development of bladder cancer (Wang et al., 2018). LncRNA DUXAP8 promotes the proliferation of bladder cancer cells by inhibiting the expression of PTEN protein (Lin et al., 2018). LncRNA ELF3-AS1 interacts with KLF8 protein molecules to form a positive feedback loop, thereby promoting the progress of bladder cancer (Guo et al., 2019).
LINC00162 is a newly discovered potential molecular target for bladder cancer, located at 21q22.3. It has been reported in skin cancer and rheumatoid arthritis (Bi et al., 2019; Piipponen et al., 2018), but its biological role and clinical significance in bladder cancer have not yet been elucidated. In this study, we observed that LINC00162 expression in bladder cancer cell lines and bladder cancer tissues was significantly upregulated. Further experiments proved that knocking down the expression of LINC00162 significantly inhibited the proliferative activity of bladder cancer cells and the ability to form monoclonal colonies in soft agar, as well as the tumor growth in nude mice, and at the same time reduced the apoptosis rate. Further, upregulating the expression of LINC00162 would produce the opposite cell phenotype change. It is suggested that LINC00162 may play a role as an oncogene in the development of bladder cancer. To clarify the molecular mechanism of LINC00162 regulating bladder cancer cell proliferation, we used CHIRP experiments to prove that LINC00162 could bind to its neighboring gene, PTTG1IP. Previously, PTTG1IP was reported to participate in the regulation of cancer development such as non-small cell lung cancer, breast cancer, glioma, and thyroid cancer through various methods such as high methylation, increasing ubiquitination, and regulating the cell cycle (Read et al., 2014; Repo et al., 2017; Tan et al., 2019; Wang et al., 2014). However, the biological function of PTTG1IP in bladder cancer has not been reported. This study is the first to explain the function of PTTG1IP in bladder cancer. RNA-FISH experiments were used to detect the subcellular localization of LINC00162. The results show that LINC00162 is mainly distributed in the nucleus, suggesting that LINC00162 may play a role in the transcription process. Through further experiments, we found that LINC00162 could regulate the expression of PTTG1IP, and these two molecules are also closely related in cell function. In order to explore the molecular mechanism of LINC00162 regulating PTTG1IP, we used RNA pull-down experiments to pull down the proteins that bind to LINC00162. And by mass spectrometry analysis, we screened one of the most abundant transcription-related proteins, THRAP3. THRAP3 is involved in the transcriptional regulation of multiple pathways as a transcriptional regulation factor. For example, it participates in the formation of the SOX9 transcription complex and negatively regulates the transcriptional activity of SOX9 (Sono et al., 2018). THRAP3 also works in concert with HELZ2 to enhance PPARG-mediated transcriptional activation (Lande-Diner et al., 2013). Through RIP experiments, we used THRAP3 antibodies to enrich nucleic acid molecules that bind to THRAP3 protein and found that both LINC00162 and PTTG1IP could bind to THRAP3 protein. Further Western blot experiments proved that THRAP3 had a positive regulation effect on the protein expression of PTTG1IP. Based on the above results, it is suggested that LINC00162 may block the pathway that THRAP3 promotes the transcription of PTTG1IP, thereby inhibiting the expression of PTTG1IP and then promoting the proliferation of bladder cancer cells. However, the specific molecular mechanism of the interaction between LINC00162, THRAP3, and PTTG1IP needs further study.
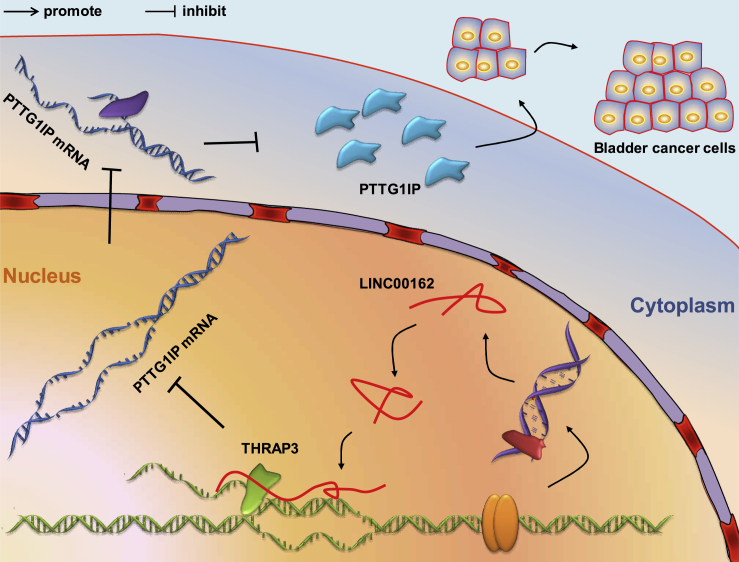
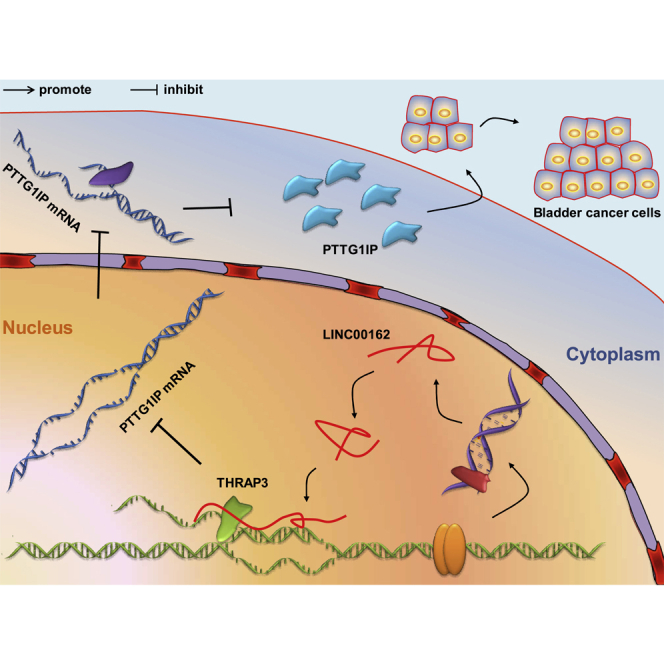
Our research shows that LINC00162 is an oncogene in the development of bladder cancer, and it inhibits PTTG1IP expression by interacting with THRAP3, thereby promoting the proliferation of bladder cancer cells (Figure 6). This study allowed us to understand the molecular function of LINC00162 more comprehensively. Moreover, we achieved a preliminary exploration of the regulatory mechanism of THRAP3 and PTTG1IP in bladder cancer. Our work provides basic support for the discovery of potential target molecules of bladder cancer.
Figure 6.
Mechanism Diagram
Schematic diagram of the molecular mechanism of LINC00162 regulating bladder cancer proliferation.
Limitations of the Study
The specific experimental mechanism on how LINC00162 and THRAP3 complex inhibit PTTG1IP expression has not been fully elucidated.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Aruo Nan (nanaruo@163.com).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession number for the Super-enhancer lncRNA microarray dataset reported in this paper is GEO: GSE159682. Other data are included in the published article and the supplemental information files and any additional information will be available from the lead contact upon request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Dapang Rao, the Second Affiliated Hospital & Yuying Children's Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325027, China, for critical technical support. This work was supported by the National Natural Science Foundation of China (NSFC81903356, NSFC81673135).
Authors Contribution
A.R.N. and H.S.H. conceived of the study. X.W., R.R.Z., and S.L.W. performed the experiments. L.P.S., M.X.K., Y.O.Y., M.Q.L., Y.T.L., B.N.S., Z.J.Z., Y.P.Y., and W.M.L. participated in statistical analysis of data, result arrangement and drawing, and manuscript writing. J.L.Y., J.Y., and D.N.L. collected clinical samples. A.R.N. and Y.F.Z. revised the paper. All authors read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: December 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101857.
Contributor Information
Haishan Huang, Email: haishan_333@163.com.
Aruo Nan, Email: nanaruo@163.com.
Supplemental Information
References
- Antoni S., Ferlay J., Soerjomataram I., Znaor A., Jemal A., Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur. Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Atianand M.K., Fitzgerald K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazrafshani M.R., Nowshadi P.A., Shirian S., Daneshbod Y., Nabipour F., Mokhtari M., Hosseini F., Dehghan S., Saeedzadeh A., Mosayebi Z. Deletion/duplication mutation screening of TP53 gene in patients with transitional cell carcinoma of urinary bladder using multiplex ligation-dependent probe amplification. Cancer Med. 2016;5:145–152. doi: 10.1002/cam4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Guo X.H., Mo B.Y., Wang M.L., Luo X.Q., Chen Y.X., Liu F., Olsen N., Pan Y.F., Zheng S.G. LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine. 2019;50:408–420. doi: 10.1016/j.ebiom.2019.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu M., Meng F., Sun B., Jin X., Jia C. The long noncoding RNA HIF1A-AS2 facilitates cisplatin resistance in bladder cancer. J. Cell. Biochem. 2019;120:243–252. doi: 10.1002/jcb.27327. [DOI] [PubMed] [Google Scholar]
- Feng F., Chen A., Huang J., Xia Q., Chen Y., Jin X. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/beta-catenin pathway axis. J. Cell. Biochem. 2018;119:9408–9418. doi: 10.1002/jcb.27257. [DOI] [PubMed] [Google Scholar]
- Guo Y., Chen D., Su X., Chen J., Li Y. The lncRNA ELF3-AS1 promotes bladder cancer progression by interaction with Kruppel-like factor 8. Biochem. Biophys. Res. Commun. 2019;508:762–768. doi: 10.1016/j.bbrc.2018.11.183. [DOI] [PubMed] [Google Scholar]
- Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. An atlas of human long non-coding RNAs with accurate 5' ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhang A., Ho T.T., Zhang Z., Zhou N., Ding X., Zhang X., Xu M., Mo Y.Y. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C., Henrique R. Epigenetic biomarkers in urological tumors: a systematic review. Cancer Lett. 2014;342:264–274. doi: 10.1016/j.canlet.2011.12.026. [DOI] [PubMed] [Google Scholar]
- Jin X., Xu X.E., Jiang Y.Z., Liu Y.R., Sun W., Guo Y.J., Ren Y.X., Zuo W.J., Hu X., Huang S.L. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci. Adv. 2019;5:eaat9820. doi: 10.1126/sciadv.aat9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R., Zhang E.B., Yin D.D., You L.H., Xu T.P., Chen W.M., Xia R., Wan L., Sun M., Wang Z.X. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L., Boyault C., Kim J.Y., Weitz C.J. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc. Natl. Acad. Sci. U S A. 2013;110:16021–16026. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang X., Wen C., Huo Z., Wang W., Zhan Q., Cheng D., Chen H., Deng X., Peng C. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol. Cancer. 2017;16:169. doi: 10.1186/s12943-017-0738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.C., Ren J.L., Wong C.W., Chan S.O., Waye M.M., Fu W.M., Zhang J.F. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/beta-catenin signaling. Oncogene. 2018;37:1445–1456. doi: 10.1038/s41388-017-0041-y. [DOI] [PubMed] [Google Scholar]
- Lin M.G., Hong Y.K., Zhang Y., Lin B.B., He X.J. Mechanism of lncRNA DUXAP8 in promoting proliferation of bladder cancer cells by regulating PTEN. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3370–3377. doi: 10.26355/eurrev_201806_15158. [DOI] [PubMed] [Google Scholar]
- Maia M.C., Grivas P., Agarwal N., Pal S.K. Circulating tumor DNA in bladder cancer: novel applications and future directions. Eur. Urol. 2018;73:541–542. doi: 10.1016/j.eururo.2017.10.011. [DOI] [PubMed] [Google Scholar]
- Misawa A., Takayama K., Urano T., Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J. Biol. Chem. 2016;291:17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales E.E., Grill S., Svatek R.S., Kaushik D., Thompson I.M., Jr., Ankerst D.P., Liss M.A. Finasteride reduces risk of bladder cancer in a large prospective screening study. Eur. Urol. 2016;69:407–410. doi: 10.1016/j.eururo.2015.08.029. [DOI] [PubMed] [Google Scholar]
- Niu Y., Ma F., Huang W., Fang S., Li M., Wei T., Guo L. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol. Cancer. 2017;16:5. doi: 10.1186/s12943-016-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel N., Couteau J., Maillet G., Gobet F., D'Aloisio F., Minier C., Pfister C. TP53 and FGFR3 gene mutation assessment in urine: pilot study for bladder cancer diagnosis. Anticancer Res. 2015;35:4915–4921. [PubMed] [Google Scholar]
- Onal B., Han U., Yilmaz S., Koybasioglu F., Altug U. The use of urinary nuclear matrix protein 22 (NMP22) as a diagnostic adjunct to urine cytology for monitoring of recurrent bladder cancer--institutional experience and review. Diagn. Cytopathol. 2015;43:307–314. doi: 10.1002/dc.23239. [DOI] [PubMed] [Google Scholar]
- Parasramka M.A., Maji S., Matsuda A., Yan I.K., Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol. Ther. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee H., Han N., Kwak S., Lee H.T., Kim J.H., Kang K., Youn B.H., Yang J.H., Jeong H.J. Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization. Nucleic Acids Res. 2018;46:11759–11775. doi: 10.1093/nar/gky923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Li H. The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol. Carcinog. 2019;58:1465–1480. doi: 10.1002/mc.23028. [DOI] [PubMed] [Google Scholar]
- Piipponen M., Heino J., Kahari V.M., Nissinen L. Long non-coding RNA PICSAR decreases adhesion and promotes migration of squamous carcinoma cells by downregulating alpha2beta1 and alpha5beta1 integrin expression. Biol. Open. 2018;7:bio037044. doi: 10.1242/bio.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbevsek P., Fasolo F., Bon C., Cimatti L., Reisser S., Carninci P., Bussi G., Zucchelli S., Plavec J., Gustincich S. Structural determinants of the SINE B2 element embedded in the long non-coding RNA activator of translation AS Uchl1. Sci. Rep. 2018;8:3189. doi: 10.1038/s41598-017-14908-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntoni M., Petrera M., Campora S., Garrone E., Defferrari C., Torrisi R., Johansson H., Bruno S., Curotto A., DeCensi A. Prognostic significance of VEGF after twenty-year follow-up in a randomized trial of fenretinide in non-muscle-invasive bladder cancer. Cancer Prev. Res. (Phila) 2016;9:437–444. doi: 10.1158/1940-6207.CAPR-15-0345. [DOI] [PubMed] [Google Scholar]
- Read M.L., Seed R.I., Fong J.C., Modasia B., Ryan G.A., Watkins R.J., Gagliano T., Smith V.E., Stratford A.L., Kwan P.K. The PTTG1-binding factor (PBF/PTTG1IP) regulates p53 activity in thyroid cells. Endocrinology. 2014;155:1222–1234. doi: 10.1210/en.2013-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repo H., Gurvits N., Loyttyniemi E., Nykanen M., Lintunen M., Karra H., Kurki S., Kuopio T., Talvinen K., Soderstrom M. PTTG1-interacting protein (PTTG1IP/PBF) predicts breast cancer survival. BMC Cancer. 2017;17:705. doi: 10.1186/s12885-017-3694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A.G., Kim J., Al-Ahmadie H., Bellmunt J., Guo G., Cherniack A.D., Hinoue T., Laird P.W., Hoadley K.A., Akbani R. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–556 e525. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roisman A., Castellano G., Navarro A., Gonzalez-Farre B., Perez-Galan P., Esteve-Codina A., Dabad M., Heath S., Gut M., Bosio M. Differential expression of long non-coding RNAs are related to proliferation and histological diversity in follicular lymphomas. Br. J. Haematol. 2019;184:373–383. doi: 10.1111/bjh.15656. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Sono T., Akiyama H., Miura S., Deng J.M., Shukunami C., Hiraki Y., Tsushima Y., Azuma Y., Behringer R.R., Matsuda S. THRAP3 interacts with and inhibits the transcriptional activity of SOX9 during chondrogenesis. J. Bone Miner Metab. 2018;36:410–419. doi: 10.1007/s00774-017-0855-2. [DOI] [PubMed] [Google Scholar]
- Tan S.K., Pastori C., Penas C., Komotar R.J., Ivan M.E., Wahlestedt C., Ayad N.G. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer. 2018;17:74. doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Zhang S., Gao H., He W., Xu M., Wu Q., Ni X., Jiang H. Hypermethylation of the PTTG1IP promoter leads to low expression in early-stage non-small cell lung cancer. Oncol. Lett. 2019;18:1278–1286. doi: 10.3892/ol.2019.10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zu Y., Zhu S., Yang Y., Huang W., Xie H., Li G. Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem. Biophys. Res. Commun. 2018;507:231–235. doi: 10.1016/j.bbrc.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Wang X.P., Deng X.L., Li L.Y. MicroRNA-584 functions as a tumor suppressor and targets PTTG1IP in glioma. Int. J. Clin. Exp. Pathol. 2014;7:8573–8582. [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Huang Z., Zhou R., Zhang J., Yu B. The prognostic value of expression of the long noncoding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) in patients with solid malignant tumors: a systematic review and meta-analysis. Med. Sci. Monit. 2018;24:5462–5472. doi: 10.12659/MSM.911687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Liao X., Chen Z., Fang Y., He A., Zhong Y., Gao Q., Xiao H., Li J., Huang W. LncRNA MALAT1 inhibits apoptosis and promotes invasion by antagonizing miR-125b in bladder cancer cells. J. Cancer. 2017;8:3803–3811. doi: 10.7150/jca.21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhang Y., Du L., Jiang X., Yan S., Duan W., Li J., Zhan Y., Wang L., Zhang S. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 2018;12:648–658. doi: 10.1002/1878-0261.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Xu Q., Kuang D., Wang Z., Lu Q., Lin Q., Wu H., Chen L. Long noncoding RNA SLNCR1 regulates nonsmall cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol. Med. Rep. 2019;20:2591–2596. doi: 10.3892/mmr.2019.10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Zhang G., Li S., Lu J., Ge Y., Wang Q., Ma G., Zhao Q., Wu D., Gong W., Du M. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer. 2018;17:87. doi: 10.1186/s12943-018-0829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang Z., Yu Q., Shen J., He W., Zhou D., Yu Q., Fan J., Gao S., Duan L. Atractylenolide II reverses the influence of lncRNA XIST/miR-30a-5p/ROR1 axis on chemo-resistance of colorectal cancer cells. J. Cell. Mol. Med. 2019;23:3151–3165. doi: 10.1111/jcmm.14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yuan W., Song J., Wang S., Gu X. LncRna CPS1-IT1 suppresses cell proliferation, invasion and metastasis in colorectal cancer. Cell Physiol. Biochem. 2017;44:567–580. doi: 10.1159/000485091. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Qian W., Wang S., Ji D., Wang Q., Li J., Peng W., Gu J., Hu T., Ji B. Analysis of lncRNA-associated ceRNA network reveals potential lncRNA biomarkers in human colon adenocarcinoma. Cell Physiol. Biochem. 2018;49:1778–1791. doi: 10.1159/000493623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the Super-enhancer lncRNA microarray dataset reported in this paper is GEO: GSE159682. Other data are included in the published article and the supplemental information files and any additional information will be available from the lead contact upon request.