Figure 4.
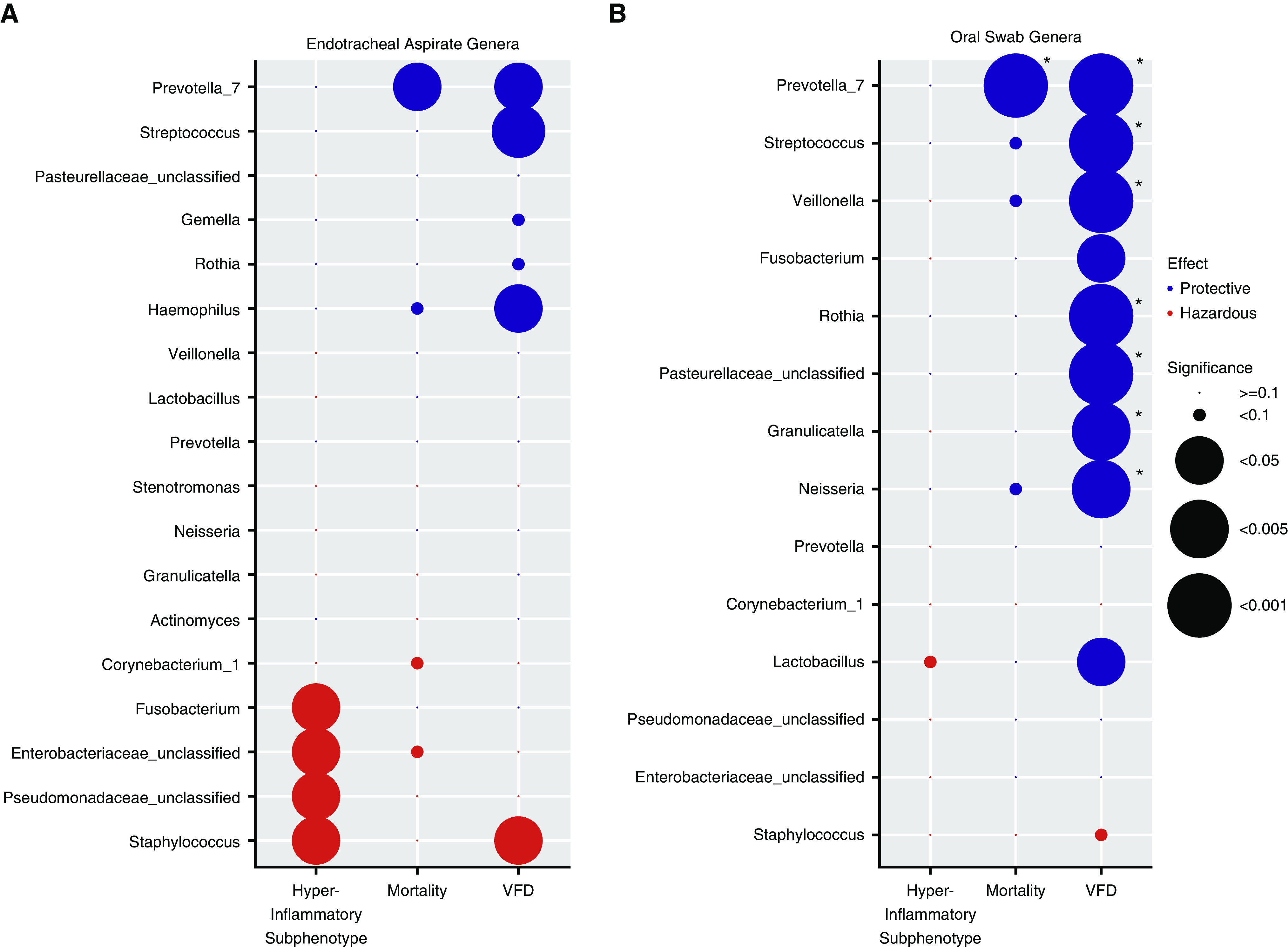
The relative abundance of individual genera is associated with clinical outcomes and host-response subphenotypes. (A) Endotracheal aspirate (ETA) genera. We examined data for associations between additive log ratio–transformed relative abundance for the top 10 genera in each cluster (total of 18 unique genera), which are shown on the y-axis with three outcome variables: hyperinflammatory subphenotype, 30-day mortality (logistic regression models), and ventilator-free days (VFDs; linear regression model). Models were adjusted for age, chronic obstructive pulmonary disease, and antibiotic exposures. In each column, the direction of the effect size of the coefficient and the statistical significance for each genus–outcome association are visually represented by color coding (with protective effects shown in blue and adverse effects shown in red) and the size of each circle, respectively. Typical pathogenic genera in ETA samples (Staphylococcus, Pseudomonadaceae, and Enterobacteriaceae) were associated with higher odds of having the hyperinflammatory subphenotype classification and fewer VFDs in the case of Staphylococcus genera, whereas typical members of the normal lung microbiome (e.g., Prevotella_7 and Streptococcus) were associated with improved outcomes. (B) Oral-swab genera. Among the 14 unique genera examined in oral swabs, a high relative abundance of typical members of the normal lung microbiome (e.g., Prevotella_7, Streptococcus, Veillonella, Rothia, etc.) was associated with improved outcomes (mainly more VFDs). Associations that remained significant after adjustment for multiple testing with the Benjamini-Hochberg method are highlighted with asterisks (*adjusted P < 0.05). In the case of Pseudomonadaceae_unclassified, Enterobacteriaceae_unclassified, and Pasterellaceae_unclassified, classification to specific genera within these families was not accomplished, and we thus used family-level descriptors for these genera.
