Figure 5. Regulatory mechanisms of GWAS loci.
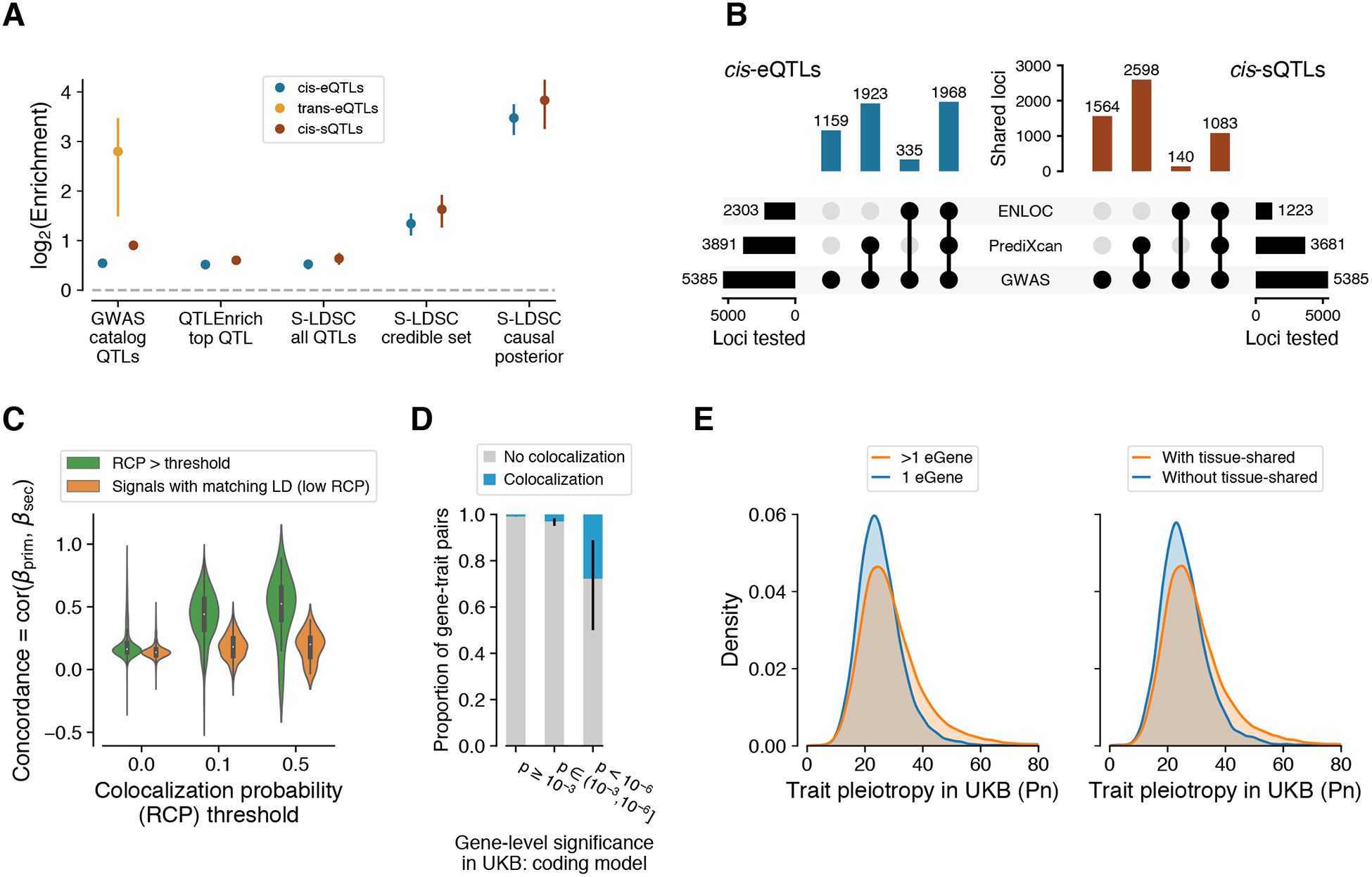
(A) GWAS enrichment of cis-eQTLs, cis-sQTLs, and trans-eQTLs measured with different approaches: enrichment calculated from GWAS summary statistics of the most significant cis-QTL per eGene/sGene with QTLEnrich and LD Score regression with all significant cis-QTLs (S-LDSC all cis-QTLs), simple QTL overlap enrichment with all GWAS catalog variants, and LD Score regression with fine-mapped cis-QTLs in the 95% credible set (S-LDSC credible set) and using posterior probability of causality as a continuous annotation (S-LDSC causal posterior). Enrichment is shown as mean and 95% CI. (B) Number of GWAS loci linked to e/sGenes through colocalization (ENLOC) and association (PrediXcan), aggregated across tissues. (C) Concordance of mediated effects among independent cis-eQTLs for the same gene, shown for different levels of regional colocalization probability (RCP (32)), which is used as a proxy for the gene’s causality. As the null, we show the concordance for LD matched genes without colocalization. (D) Proportion of colocalized cis-eQTLs with a matching phenotype for genes with different level of rare variant trait association in the UK Biobank (UKB). (E) Horizontal GWAS trait pleiotropy score distribution for cis-eQTLs that regulate multiple vs. a single gene (left), and for cis-eQTLs that are tissue-shared vs. specific.
