Figure 1.
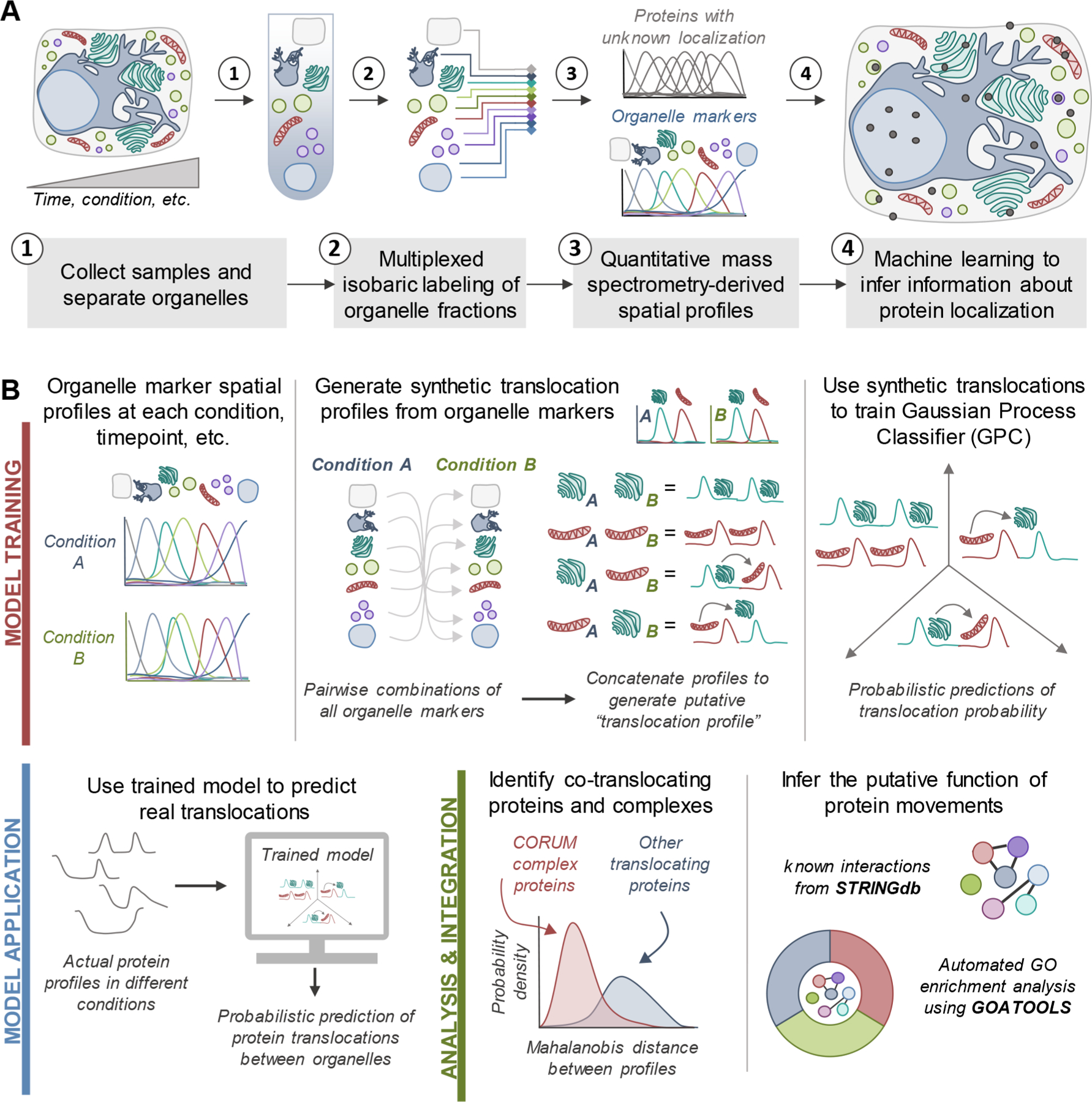
Workflows for spatial proteomics and the TRANPIRE pipeline for studying protein localization and movement. (A) General workflow for assessing the subcellular distribution of cellular proteins using organelle fractionation-based spatial proteomics. Most commonly, these approaches combine multiplexed isobaric labeling and quantitative mass spectrometry with machine learning to discern information regarding protein localization. Organelle marker proteins are used to inform machine learning-enabled classification. (B) Data processing and analysis pipeline for TRANSPIRE, a computational method leveraging a Gaussian process classifier (GPC) to characterize protein movements between organelles. By training the classifier with synthetic translocation profiles derived from different combinations of organelle markers, this classifier can detect and score the probability of protein translocation events. The output of the classifier is further combined with Mahalanobis distance analyses to identify co-translocating proteins and protein complexes. Integration of this analysis with known interactions and gene ontology enrichment can help reveal the putative function of these changes in protein distribution.
