Figure 2.
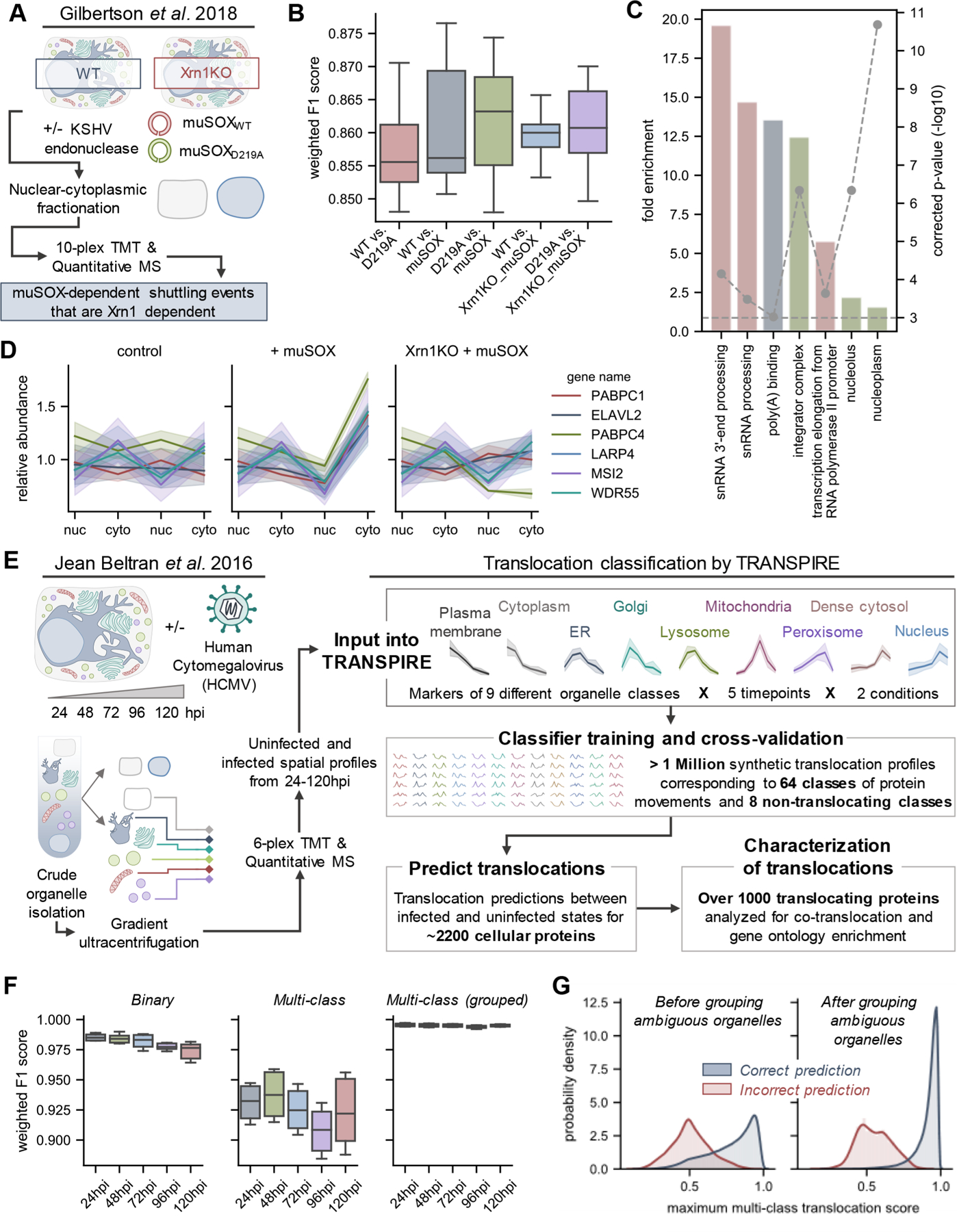
Assessing the reliability of TRASPIRE classification for predicting protein translocations in the context of viral infection. (A) Experimental workflow from Gilbertson et al.,53 a study focused on understanding nuclear-cytoplasmic shuttling events upon KSHV-induced cellular mRNA decay (induced by transfection of the KSHV endonuclease muSOX or its catalytically inactive counterpart muSOX D219A). (B) Boxplots of weighted F1 scores describing classifier performance across five balanced training folds and three biological replicates for each experimental condition. (C) Gene ontology enrichment on proteins predicted to translocate by TRANSPIRE point to RNA binding proteins, in agreement with the results of the original manuscript. (D) Selected profiles of proteins predicted to translocate by TRANSPIRE in an Xrn1-dependent manner. Solid lines and shaded areas represent the mean and standard deviation, respectively, of protein profiles across the three biological replicates reported in the study. (E) Experimental workflow for the HCMV spatial proteomics study that was subsequently analyzed by TRANSPIRE. Using 6-plex TMT labeling, Jean Beltran et al. generated spatial profiles of proteins in uninfected and (HCMV)-infected cells at 24, 48, 72, 96, and 120 hpi. The curated organelle markers defined in the original study were used to generate synthetic translocation profiles from all pairwise combinations of organelle markers. Equal subsets of profiles corresponding to each combination of markers were then used to train the classifier and performance was validated on a held-out subset of test data. Following training, the classifier was then applied to predict translocations within the data set and high confidence predictions were further characterized by integrating information regarding known protein interactions and gene ontology enrichment analysis. (F) Boxplots of weighted F1 scores describing performance on the held-out test data set across all time points of infection at both binary (e.g., translocating versus not translocating) and multiclass levels, as well as for classifier predictions after grouping ambiguous organelles. Ambiguous organelle groups were: plasma membrane/cytoplasm, ER/Golgi/lysosome, and dense cytosol/nucleus. (G) Classifier score distributions for correct and incorrect classifier predictions before and after grouping ambiguous organelles. Note that these scores refer to multiclass translocation assignments rather than the binary translocation scores discussed later in the manuscript.
