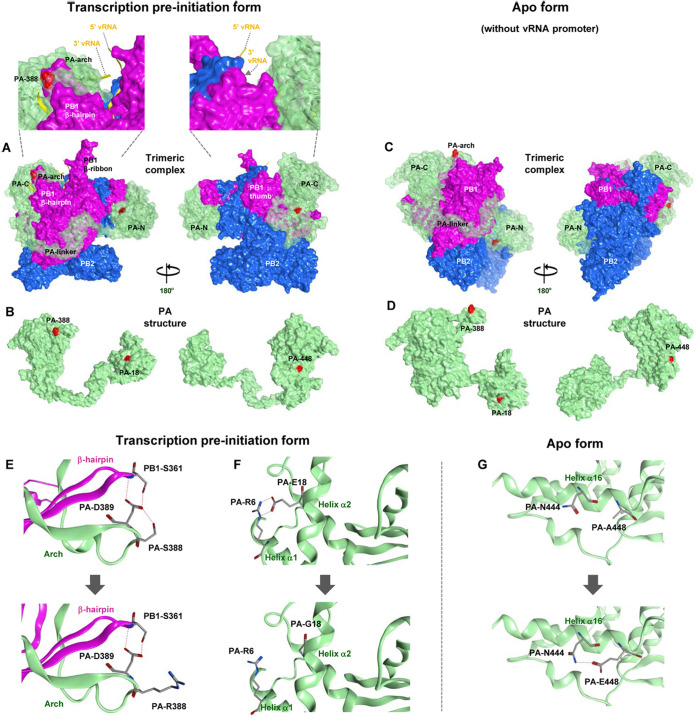
FIG 9.
Structural model of the clade 2.2.1 polymerase complex. Shown is a structural model of the D1/PA heterotrimeric polymerase complex. (A and C) Transparent surface diagrams of the D1/PA polymerase complex composed of PB2 (blue), PB1 (pink), and PA (green) with the A448E, S388R, and E18G mutations (red). (A) The transcription preinitiation form (PDB accession number 6RR7 as the modeling template), where the polymerase complex binds to the vRNA promoter (yellow). A close-up of the polymerase complex around the vRNA promoter is also shown. (C) The apo form (without the vRNA promoter) (PDB accession number 6QPF as the modeling template). (B and D) Surface views of the PA structure in the transcription preinitiation form (B) and the apo form (D). The left and right PA structures differ by 180° in orientation. (E to G) Polymerase structure without (top) and with (bottom) the indicated PA mutations in the D1/PA heterotrimeric polymerase complex with vRNA. (E) Close-up of residue 388 in the D1/PA polymerase complex. (F) Close-up of residue 18 in the D1/PA polymerase complex. (G) Close-up of residue 448 in the D1/PA polymerase complex. Potential interactions between residues are represented by broken lines.