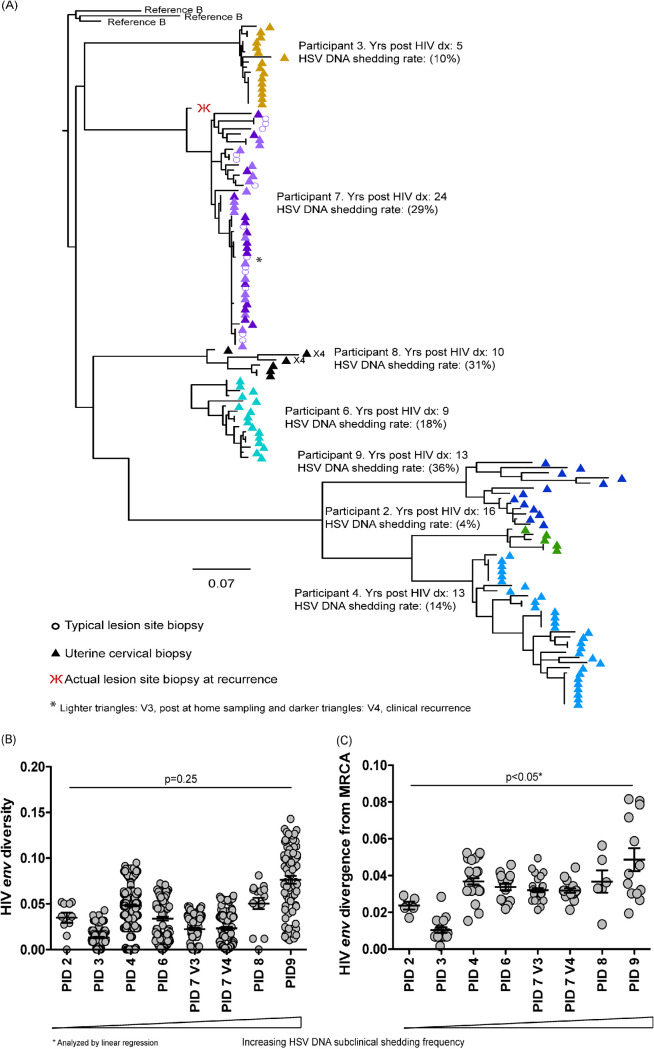
FIG 4.
Higher HSV shedding is associated with increased divergence from the most common recent ancestor (MRCA). Nucleic acids from typical lesion site (TLSB) and uterine cervical biopsy specimens at V3 and for one clinical recurrence (at V4) were submitted for single genome amplification (SGA) of HIV envC2-V5 and aligned in Geneious. (A) A phylogenetic tree displaying sequences from all seven participants with successful HIV SGA were rooted to Reference B sequences. Most participants’ sequences were generated from their cervical tissues at V3 (participants 2, 3, 4, 6, 8, and 9) and are represented as different colored triangles by participant. Participant 7 has sequences amplified from her TLSB and cervical tissues from V3 represented by light colored purple triangles for her cervical sequences and by open light purple circles for her TLSB sequence; her clinical recurrence cervical sequences are represented as the darker purple triangles with one HIV DNA sequence from her actual lesion site biopsy specimen (ALB) (red psi symbol). Sequences were rooted using representative sequences for subtype B from the GenBank database (clade B: B.US.JRFL, B.US.90.WEAU160, B.FR.83.HXB2). The scale bar (horizontal line) indicates the number of substitutions per site. (B and C) Phylogenetic pairwise distance analyses were generated in DIVEIN (25) and diversity (B) and divergence (C) data were visualized using GraphPad Prism (GraphPad).