FIGURE 7.
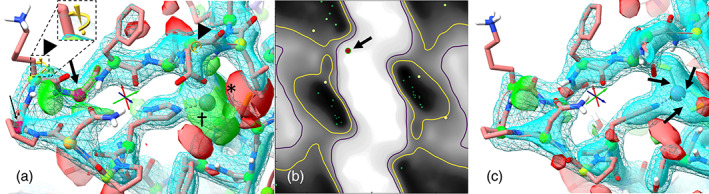
Typical scenes from an ISOLDE session, rebuilding a problematic loop in an x‐ray crystallographic model (PDBid: 7bq7). (a) Model as loaded from the wwPDB. Maps are calculated from the experimental structure factors and are updated on the fly in response to coordinate changes. Cyan wireframe/surface: electron density map at two different contours; green/red surface: difference map at +/− 3σ. Cryo‐EM sessions differ only in that the provided map(s) do not change over time. Live markup includes questionable rotamers (inset; marker grows and shades from yellow to red with decreasing probability) and backbone Ramachandran status (colored balls overlaid on alpha carbon atoms, shading from green through yellow to red with decreasing probability). This scene contains two Ramachandran outliers (arrows), two marginal rotamers (arrowheads), and a cysteine sidechain badly out of density as shown by the difference map (*/† show current and expected positions respectively). (b) ISOLDE's interactive Ramachandran plot, showing the glycine outlier selected in (a). This plot updates in real‐time. Hovering over a point identifies it, and clicking displays the corresponding residue in the main view. (c) Corrected model. Interactive distance restraints were applied to maintain the correct zinc‐sulfur distances in the Cys3HisZn coordination site at right (arrows)
