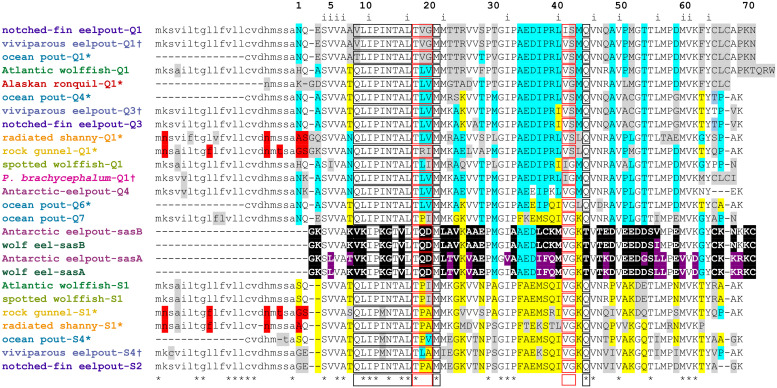
Fig 2. Alignment of a representative selection of known type III AFP sequences.
Sequences are named using the common name for each species except for P. brachycephalum. Names are colored as in Fig 1 with an asterisk indicating the sequences were obtained by PCR in this study (S3 Table) and with a dagger for those assembled from the SRA database (S4 Table). They are numbered consecutively by species within the QAE (Q) or SP (S) group as they appear in S3 Fig. Residues conserved in all isoforms (sasA and B) are indicated with asterisks, variations characteristic of the QAE and SP groups are highlighted cyan and yellow respectively, with other variations in grey. Shared differences in the signal peptides of radiated shanny and rock gunnel are highlighted red. Black and red boxes show residues on the pyramidal- and prism-plane binding surfaces respectively [79]. Inward-facing residues (i) are indicated along the top. Residues unique to SAS are highlighted black with other differences within this group highlighted purple. Signal peptides are in lower case font. Dashes indicate gaps (internal) or incomplete sequence (at termini). The complete protein and nucleotide alignments are shown in S3 and S4 Figs, respectively.