Extended Data Fig. 4. High-order enhancer component assemblies mediated by differential TF-TF and TF-enhancer interactions correspond with endocrine resistance-associated enhancer reprogramming.
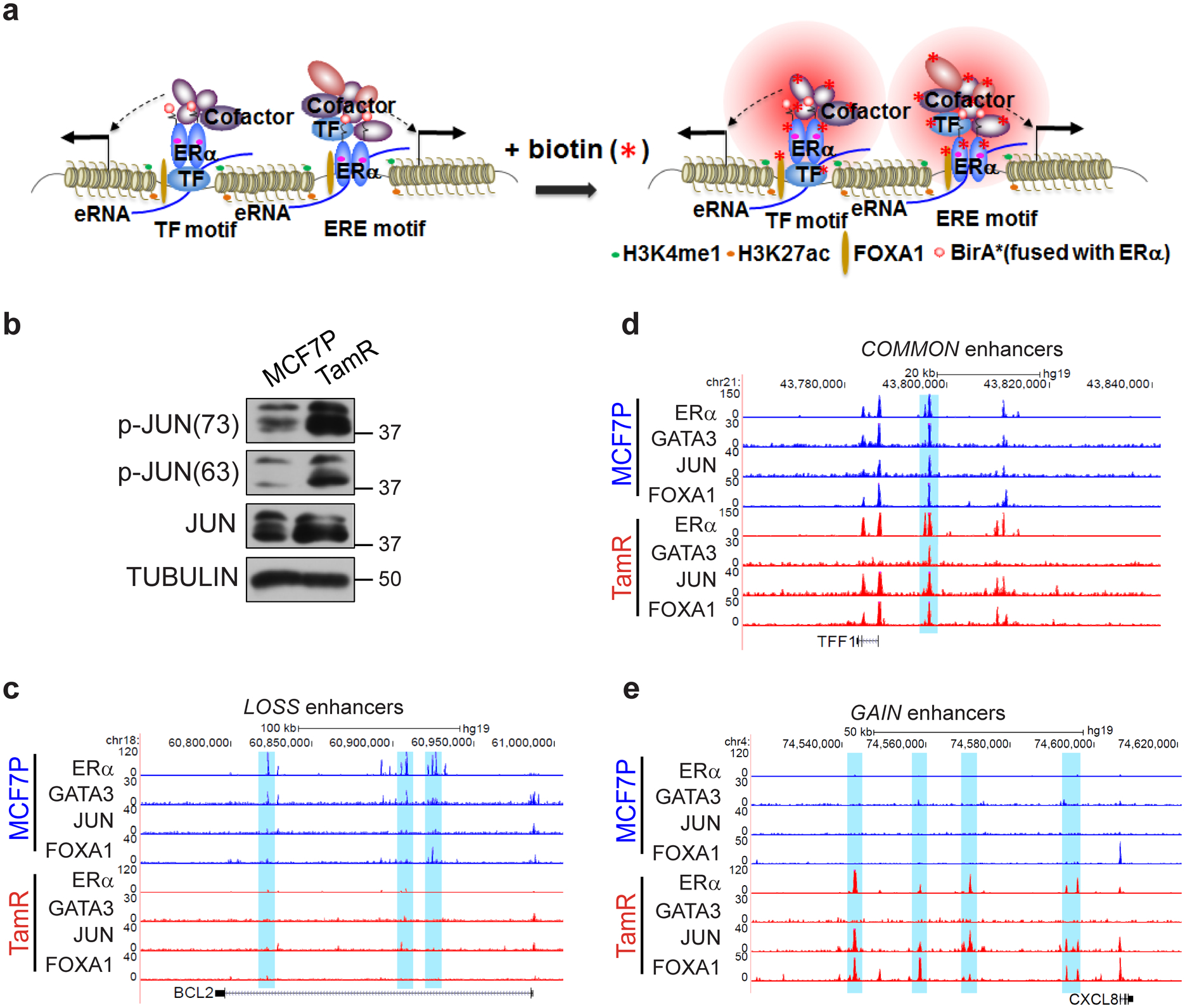
a, Schematic diagram of BioID (in vivo proximity-dependent biotin identification) approach for identification of ERα-interacting nuclear proteins including both TFs and other transcriptional cofactors in alive cells. This technology was used to explore the ERα-interacting (or in the close proximity) enhancer components in either endocrine-sensitive or -resistant cellular context.
b, Western blot analyses of total JUN or phosphorylated JUN protein levels in MCF7P and TamR cells. Tubulin was used as a loading control.
c-e, Genome browser snap images of ChIP-seq data showing the co-binding of GATA3, JUN, FOXA1 and ERα at the LOSS enhancer regions near BCL2 gene (c), the COMMON enhancer regions near TFF1 gene (d), and GAIN enhancer regions near CXCL8 gene (e) in both MCF7P and TamR cell lines.
Immunoblots are representative of two independent experiments. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 4.
