Fig. 5. GATA3 is required for maintenance of LOSS enhancers and expression of epithelial makers.
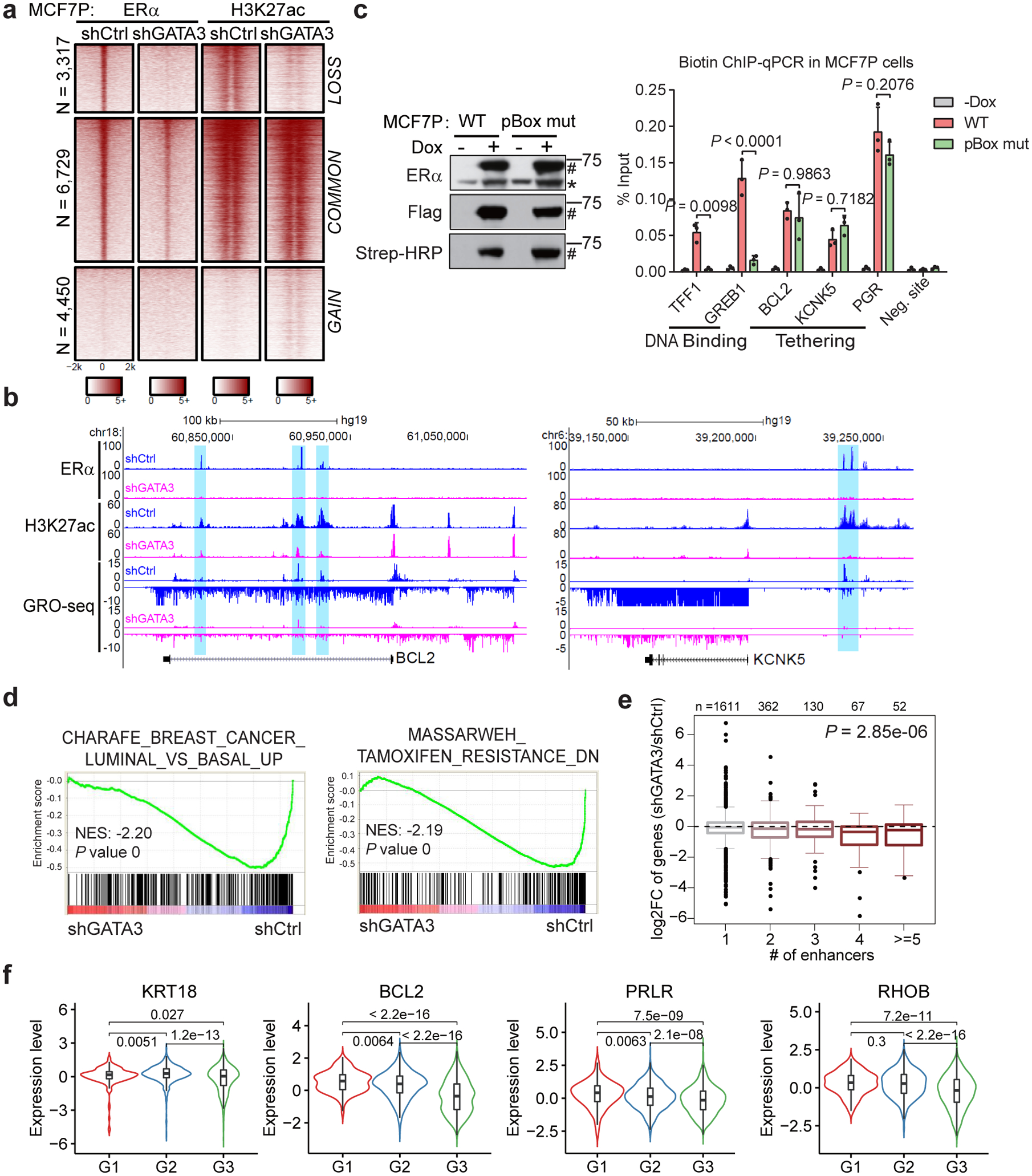
a, Heatmaps of ERα and H3K27ac ChIP-seq demonstrating that knockdown of GATA3 in MCF7P cells results in enhancer inactivation for the LOSS enhancers.
b, Genome browser views of GRO-seq data and ChIP-seq data at BCL2 and KCNK5 gene loci demonstrating enhancer inactivation and downregulation of gene expression upon depletion of GATA3 in MCF7P cells.
c, Western blots (left) showing doxycycline-induction and in vivo biotinylation of BLRP-tagged ERα WT and pBox mutant in MCF7P cells. * and # indicate endogenous and tagged exogenous ERα respectively. ChIP-qPCR showing that ERα binding on the classical ERE-containing enhancers at TFF1 and GREB1 loci was abolished by the mutation in ERα’s pBox DNA-binding domain. However, the recruitment of ERα to LOSS enhancers (on BCL2, KCNK5 and PGR) is not affected by the mutation. Statistics: n = 3 independent experiments, mean ± s.d., two-sided t-tests.
d, GSEA analyses on RNA-seq data for MCF7P cells treated with shCtrl or shGATA3. The nominal P values were determined by empirical gene-based permutation test.
e, Integration of RNA-seq and ChIP-seq data to correlate gene regulation effects by GATA3 knockdown and GATA3-bound LOSS enhancers in MCF7P cells. Box plots showing GATA3 knockdown effects on these genes stratified by the numbers of nearest LOSS enhancers within 200 kb from the TSS site of each gene. P value was determined by ANOVA analysis.
f, Negative correlation between the average gene expression levels of validated GATA3 direct targets and tumor grades (G1, G2 and G3). METABRIC dataset were used in the analyses. n=169, 767 and 951 for G1, G2 and G3 grade samples, respectively. P values were calculated by two-sided Wilcoxon rank-sum test.
For the box plots in e and f, the lower and upper hinges correspond to the first and third quartiles, and the midline represents the median. The upper and lower whiskers extend from the hinge up to 1.5 * IQR (inter-quartile range). Outlier points are indicated if they extend beyond this range.
Immunoblots are representative of two independent experiments. Unprocessed immunoblots are shown in Source Data Fig. 5. Statistical source data are available in Statistical Source Data Fig. 5.
