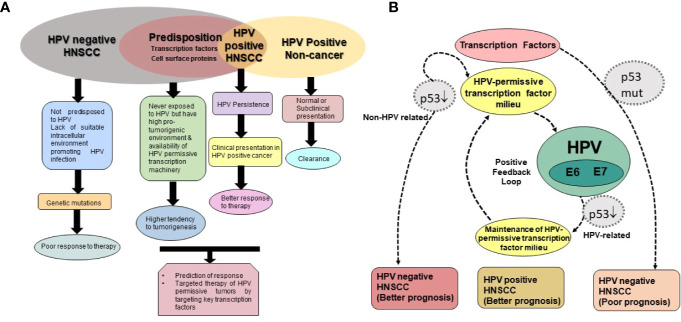
Figure 6.
Transcription factor-based classification of HPV positive and HPV negative HNSCC. (A) Schematic representation of molecular sub-types of HNSCC. HPV negative HNSCC collectively represent tumors with availability of HPV-permissive transcription factors that are never exposed to HPV infection in the lifetime and HPV negative tumors, which are driven by genetic mutations in p53 or other such genes. The later represent tumors with poorer prognosis. Further, not all individuals that encounter HPV in H&N region develop cancer. HPV positivity is reported in oral rinse of normal individual. HPV-permissive transcription factor cocktail if present is expected to direct treatment outcome similar to the tumor-tissue with HPV-driven transcription profile. In the absence of HPV infection, such cases go unnoticed and clubbed with HPV negative HNSCC and could be effectively treated with less aggressive treatment regimen and likely to have a better treatment outcome. (B) Expression of viral oncogenes under the influence of HPV-permissive transcription factor milieu lead to functional inactivation of p53 and feeds to a positive feedback look that maintains the HPV-permissive transcription factor milieu. On the other hand, HPV negative tumors, particularly with mutations in p53, in consort with other procarcinogenic transcription factors like STAT3 drive tumors that show poorer response to anti-cancer therapies.