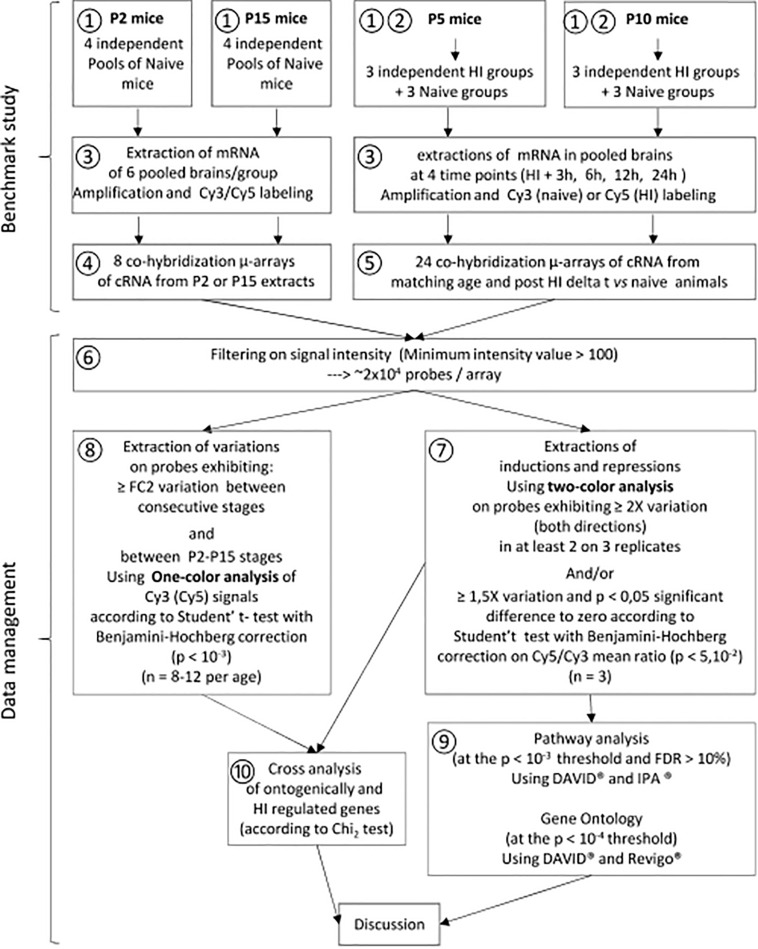
FIGURE 1.
Transcriptome study experimental schedule. Spontaneous development ① and effects of HI ② were studied in parallel. Cy3 signals were used for development study at P5 and P10. ③ mRNA extraction amplification and labeling. ④ Co-hybridization of Cy3 labeled P2 and Cy5-P15 samples. ⑤ Co-hybridization of Cy3-naïve and Cy5-HI samples at P5 or P10. ⑥ Signal filtering on signal quality and basal expression at the same thresholds. ⑦ Extraction of HI effects at P5 or P10 using a GeneSpring® two-color procedure. ⑧ Extraction of ontogenetic variations between P2, P5, P10, and P15 using a GeneSpring® one-color procedure. ⑨ Biostatistical analyses of HI effects at P5 or P10 utilizing DAVID®, Revigo®, and IPA®. ⑩ Identification of convergences between spontaneous development and effects of HI at P5 or P10 using Excel and GraphPad. cRNA: Cy3 or Cy5 labeled complementary RNA from retro-transcribed DNA obtained from extracted mRNA.