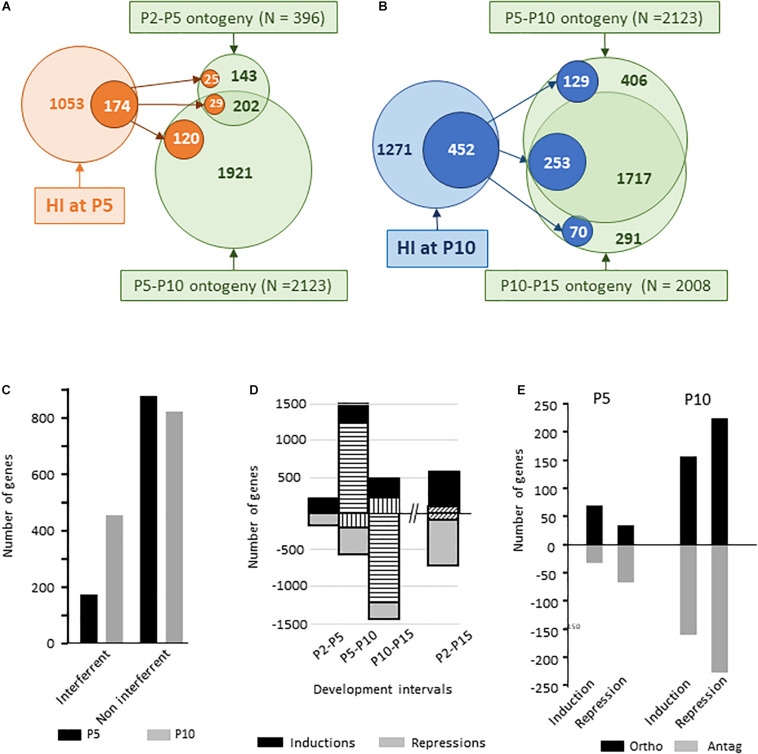
FIGURE 9.
Ontogenic regulations of gene expression in dissected hemispheres over the P2–P5, P5–P10, P10–P15, and the entire P2–P15 periods. (A) Schematic representation of HI-evoked gene expression at P5 co-incidence with previous (P2–P5) or subsequent (P5–P10) ontogenic evolution. (B) Schematic representation of HI-evoked gene expression at P10 co-incidence with previous (P5–P10) or subsequent (P10–P15) ontogenic evolution. (C) Number of genes affected by HI at P5 (black histograms) or P10 (gray histograms) also affected along with development (co-incident) or not. HI effect on genes with spontaneous development change appeared significantly enhanced in P10 mice (χ2 = 354, df; 1, p < 0.0001). (D) Number of genes induced (positive values) and repressed (negative values) over the different time intervals. Horizontal hatched histograms indicate the number of genes exhibiting P5–P10 increased expression followed by P10–P15 repression. Vertical hatched histograms indicate the number of genes exhibiting P5–P10 repression followed by P10–P15 increased expression. Oblique hatching indicates gene numbers having undergone opposite regulation in the intermediate intervals and P2–P15 net variation. (E) Distribution of HI effect on genes with spontaneous development change according to the direction of regulation; in the direction of (ortho) or opposite (antag) to developmental direction. Distribution were statistically different at P5 and P10 (χ2 = 23.8, df 3, p < 0.0001).