Abstract
Liquid-liquid phase separation (LLPS) has emerged as a central player in the assembly of membraneless compartments termed biomolecular condensates. These compartments are dynamic structures that can condense or dissolve under specific conditions to regulate molecular functions. Such properties allow biomolecular condensates to rapidly respond to changing endogenous or environmental conditions. Here, we review emerging roles for LLPS within the nuclear space, with a specific emphasis on genome organization, expression and repair. Our review highlights the emerging notion that biomolecular condensates regulate the sequential engagement of molecules in multistep biological processes.
Subject terms: Nuclear organization, Epigenetics, DNA damage and repair, Chromatin, Gene expression
Laflamme and Mekhail discuss emerging nuclear roles for LLPS in genome organization, gene expression and DNA repair, highlighting the emerging notion that biomolecular condensates regulate the sequential engagement of molecules in multistep biological processes.
Introduction
Eukaryotic cells are composed of a variety of organelles, which are typically enclosed by a membrane composed of a lipid bilayer. The main function of these membrane-bound organelles is to organize and compartmentalize the cell and to favour certain types of molecular reactions. This is in part achieved by increasing the concentrations of specific factors and by sequestering and storing other molecules. One such organelle is the nucleus, which is delimited by a double lipid bilayer called the nuclear envelope. The transport of molecules from the cytoplasm to the nucleus occurs through nuclear pore complexes that are embedded within the nuclear envelope.
In addition to membrane-bound organelles, cells contain a variety of membraneless compartments. Increasing evidence suggests that LLPS underlies the formation of such biomolecular condensates1–3. Phase separation is defined as the process by which macromolecules condense into a liquid-like dense phase, demixing from a diluted surrounding environment2,4. LLPS allows for the enrichment of specific factors while excluding others from the condensates, thereby creating a unique environment that either favours or restricts certain biochemical reactions5. LLPS drives the formation of multiple membraneless compartments, including but not limited to P granules, the nucleolus, facultative heterochromatin, transcriptional complexes and DNA repair centres6–14. These phase-separated compartments are dynamic, and the processes mediating their formation and dissolution are tightly regulated. Indeed, aberrant transition in phase separation from a liquid-like state to a more solid-like state has been linked to various human illnesses, including neurodegenerative diseases and cancer (reviewed elsewhere4,15–18).
In this review, we provide an overview of the criteria used to define LLPS and the mechanisms regulating this process. We also discuss recent LLPS literature with a focus on genome organization, expression and repair. Overall, we argue for a complex interconnection between different phase-separated intranuclear compartments.
LLPS characteristics and criteria
Membraneless organelles that form through the process of LLPS are composed of macromolecules, such as proteins and nucleic acids, which self-organize into a dynamic network of multivalent interactions. The formation of this dynamic network relies on forces promoted by weak intra- and intermolecular interactions including a combination of hydrophobic, electrostatic, cation-pi and pi-pi contacts19–21. These weak interactions allow for the rapid internal reorganization of the biomolecular condensates. The specific interactions between macromolecules within the condensates enable the formation of distinct and specialized compartments5.
Multivalency, or the presence of multiple interacting domains or elements in macromolecules, is also a critical factor mediating LLPS2,22,23. This multivalency can be achieved through the presence of repeated structured domains within proteins1,21. In addition, due to its capacity to form scaffolds and connect its multiple binding partners, RNA is an especially central player in phase separation24–27. Multivalency can also be driven by the intrinsically disordered regions (IDRs) of proteins13,28,29. IDRs do not adopt a stable three-dimensional structure, allowing IDR-containing proteins to serve as scaffolds that interact with short and flexible interacting motifs30,31. Of note, IDR mutations are frequently observed in diseases associated with LLPS dysregulation. These observations highlight the importance of the precise and dynamic regulation of biomolecular condensates. This regulation can be achieved, at least in part, via post-translational modifications (PTMs) of condensate components, which can greatly modify the charge and the conformational space of IDRs32–35. However, it is also important to note that IDRs are not always drivers of LLPS, and that IDRs can even inhibit LLPS in specific contexts29,36.
The notion that membraneless cellular organelles can form through LLPS is not a new concept. Indeed, in 1946, after studying the effect of temperature variation on the size of the nucleolus, Lars Enrenberg stated that “the nucleolus is a separated phase out of a saturated solution”37. More recently, the nucleolus, along with other major nuclear compartments, has often served as a model to study LLPS processes within the nucleus7,38,39. These studies have highlighted a number of specific criteria for defining LLPS both in vitro and in vivo (reviewed elsewhere1,3,40,41). First, due to surface tension, the shape of a biomolecular condensate is most often spherical40. Importantly, this spherical shape can be deformed when a physical force is applied or when biomolecular condensates flatten against a surface, a phenomenon known as wetting13. Second, biomolecular condensates often fuse upon touching before relaxing back into a spherical shape40. Third, biomolecular condensates contain highly dynamic molecules that exchange both within the condensate and with the surrounding environment. This rapid rearrangement of molecules in biomolecular condensates can be assessed by fluorescence recovery after photobleaching (FRAP)1. Fourth, biomolecular condensates assemble only after its components reach a saturation concentration (Csat)40. Moreover, the size of the condensate is proportional to the concentrations of its components. Fifth, the movement of components across the phase separation boundary is energetically unfavourable due to the presence of cohesive interactions within the condensate. This results in a decreased diffusion rate for molecules moving across a phase boundary3. Finally, treatment with the aliphatic alcohol 1,6-hexanediol can be used to assess the involvement of weak hydrophobic interactions during the formation of biomolecular condensates13.
Next, we will focus our discussion on three types of membraneless compartments suggested to form through LLPS inside of the nucleus. We will provide evidence for and against the involvement of phase separation in the formation of these compartments, as well as the key factors implicated in the dynamic regulation of these structures.
LLPS in heterochromatin and genome organization
In eukaryotic cells, DNA is packaged and wrapped around an octameric complex of histone proteins. These structures, called nucleosomes, represent the first level of DNA compaction and are often referred to as “beads on a string”42. These nucleosomes are then further organized and compacted to various degrees to form chromatin fibres. Heterochromatin, or silent chromatin, is critical for eukaryotic genome regulation and organization. For instance, heterochromatin formation regulates the activity of mobile genetic elements and supports the sequestration and stabilization of repetitive DNA sequences43–47. A key feature of heterochromatin is histone H3 trimethylated on lysine 9 (H3K9Me3), which is deposited by the histone methyltransferase Suv39h in mammals48. The spreading of this epigenetic mark along chromatin allows for the binding of the highly conserved heterochromatin protein 1 (HP1)49. HP1 proteins associate with H3K9Me3 via their N-terminal chromodomain and dimerize through their C-terminal chromoshadow domain50–52. Together, HP1 proteins form a platform and allow the recruitment of additional heterochromatin factors that mediate chromatin compaction and transcriptional silencing.
Recently, HP1-mediated heterochromatin formation was suggested to involve LLPS (Fig. 1a)8,9,53. Indeed, HP1 proteins purified from Drosophila melanogaster, Homo sapiens and Schizosaccharomyces pombe all undergo phase separation in vitro. Specifically, human HP1α proteins require either the phosphorylation of their N-terminus or the presence of DNA for phase separation in vitro9. Similarly, liquid droplet formation by the S. pombe HP1 protein Swi6 also requires the presence of either DNA or nucleosomes53. Most in vivo experimental observations of HP1 are also consistent with the process of LLPS. For example, Drosophila HP1a exhibits liquid-like properties in early embryos, as GFP-HP1a foci are spherical and frequently fuse. More importantly, reductions in the diffusion rate of HP1a at the heterochromatin–euchromatin border reveal the presence of a phase boundary8. Mechanistically, the binding of the S. pombe HP1 protein Swi6 to nucleosomes was shown to result in dynamic conformational changes and destabilization of the nucleosome. Such changes result in the exposure of buried nucleosomal regions, providing additional opportunities for multivalent interactions between nucleosomes and promoting LLPS53.
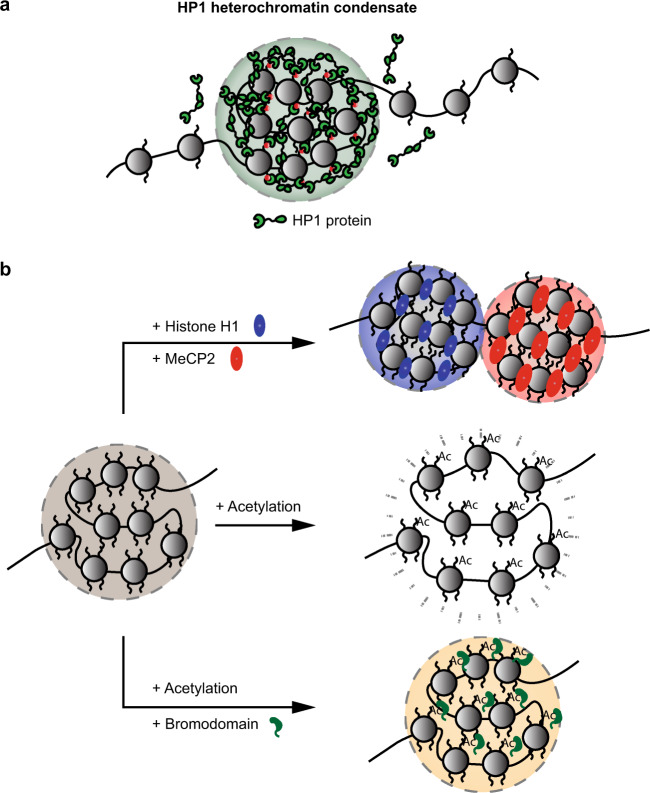
Fig. 1. LLPS in heterochromatin and genome organization.
a HP1 proteins associate with H3K9Me3 marks (red), oligomerize, and mediate the LLPS of heterochromatin. b Factors controlling the LLPS of nucleosome arrays. Biomolecular condensates formed by chromatin are modulated by several factors. Addition of the linker histone H1 (blue) and MeCP2 (red) proteins results in the formation of distinct and immiscible condensates. Acetylation of histone tails leads to the dissolution of the condensate, resulting in a more dispersed arrangement of the nucleosomes. Chromatin condensates can be reassembled upon the addition of bromodomain-containing proteins (green).
However, not all observations of heterochromatin compartments are compatible with HP1-driven phase separation. First, FRAP experiments indicated the presence of an immobilized fraction of Drosophila HP1a proteins in the late stages of developing embryos8. The immobilization of HP1 on chromatin suggests that the dynamic organization of heterochromatin through LLPS could reorganize into a more static structure during development or under certain conditions. Furthermore, studies in mouse embryonic fibroblasts indicated that HP1α organizes heterochromatin into a collapsed polymer globule without LLPS hallmarks54. Future work should aim to understand how heterochromatin-related phase separation may differ across developmental stages and organisms. It will be important to identify the factors and mechanisms driving the physiological transition of heterochromatin from a liquid-like state to other states. Such information could also shed light on pathological phase transitions observed in the context of cancer or neurological disorders.
Beyond heterochromatin, a more general capacity of chromatin to undergo LLPS is emerging. Purification of recombinant dodecameric nucleosome arrays incubated at physiological salt concentrations revealed that LLPS is in fact an intrinsic property of chromatin and can be modulated by several factors (Fig. 1b)55. The addition of linker histone H1 proteins resulted in denser and less dynamic chromatin droplets, whereas histone acetylation triggered the dissolution of these droplets. The addition of bromodomain-containing proteins such as BRD4, which binds to histone tails with acetylated lysines, restored the LLPS of acetylated chromatin56. In addition, distinct chromatin phases that are composed of acetylated and non-acetylated chromatin can coexist without coalescing55. Other factors, such as methyl-CpG-binding protein 2 (MeCP2), which selectively binds methylated CpG dinucleotides, can also induce LLPS of nucleosomal arrays. Of interest, MeCP2 competes with linker histone H1 and forms distinct heterochromatin condensates that coexist but do not mix (Fig. 1b)57. The LLPS of nucleosome arrays likely acts on a shorter length scale within chromosomes. This might in turn contribute to the dynamic compartmentalization of the genome via the formation of relatively granular chromatin subdomains with different degrees of compaction. Overall, these studies indicate that the intrinsic capacity of chromatin in general, or heterochromatin in particular, to undergo LLPS can be actively modulated by histone or DNA modifications as well as chromatin binding factors. Importantly, chromatin regulation constitutes only one approach that LLPS can employ to control gene expression.
LLPS in gene expression
Eukaryotic gene expression is regulated by an array of transcription factors and coactivators that bind to and connect promoters with enhancers. Transcription factors enable the recruitment of RNA polymerase II (RNA Pol II) to promoter regions to activate transcription58. The clustering of multiple enhancers in three-dimensional space can form super-enhancers where the transcription machinery components become highly enriched. Such super-enhancers can efficiently boost and/or coordinate gene expression at proximal genes and play a pivotal role in determining cell identity during development59,60.
The function of super-enhancers may also be shaped by LLPS10,61. Consistent with this notion, transcription factors and coactivators commonly contain IDRs that can compartmentalize and concentrate factors necessary for transcription (Fig. 2)62–68. For example, endogenous levels of the transcriptional coactivators BRD4 and a component of the Mediator complex (MED1) are enriched at super-enhancers62,63. The structures formed by these coactivators show liquid-like properties in cells. These properties include frequent fusion events, dynamic internal rearrangements of the components as evidenced by FRAP and dissolution of the foci upon 1,6-hexanediol treatment62,64. In vitro, many purified transcription factors and coactivators containing IDRs organize into phase-separated droplets that are capable of mixing with MED1 droplets63. Recently, the ability of BRD4 to undergo LLPS was shown to be dependent on the expression of specific isoforms and on phosphorylation68.
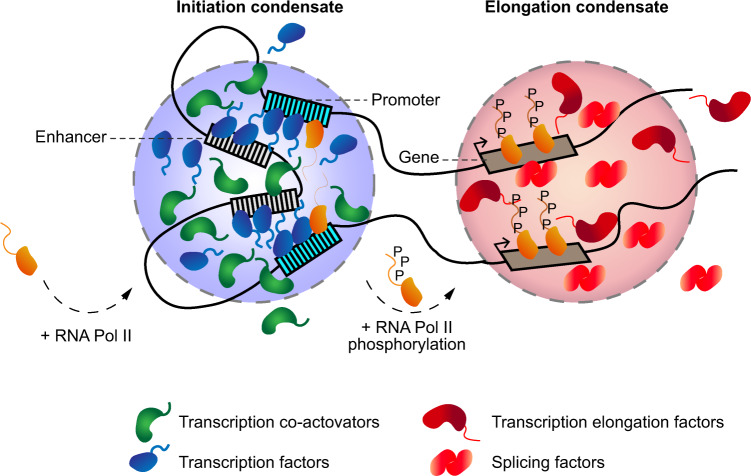
Fig. 2. LLPS in transcriptional regulation.
Biomolecular condensates composed of transcription factors and coactivators at super-enhancers and promoter sequences recruit RNA Pol II. Phosphorylation of RNA Pol II mediates its transition from the initiation condensate to the elongation condensate, which is composed of transcriptional elongation and splicing factors.
Additional factors, including members of the FUS, EWS and TAF15 (FET) family of RNA and DNA binding proteins, contain IDRs and undergo LLPS in vitro65,69,70. Proteins of the FET family also exhibit LLPS properties in vivo when tethered to artificial LacO arrays or endogenous microsatellite repeats. FET proteins form spherical and highly dynamic compartments that are capable of selectively enriching specific factors, including RNA Pol II. Interestingly, the concentration of FET proteins at the LacO site significantly exceeded the number of binding sites, suggesting the existence of cooperative binding65. Recent evidence also suggests that transcriptional control of the Hippo pathway, which is involved in various aspects of cell proliferation and differentiation71, is regulated by LLPS66,67. Under osmotic stress, YAP and TAZ transcriptional coactivators exhibit liquid-like properties in vitro and in vivo. Similar to other transcriptional coactivators, YAP and TAZ nuclear condensates co-localize with many other transcription mediators, including BRD4, MED1 and RNA Pol II66,67. Taken together, these studies suggest that LLPS at super-enhancers enables the recruitment and concentration of transcription regulators that interact through their IDRs to enrich RNA Pol II and control gene expression.
In addition to concentrating the factors involved in transcription initiation, recent evidence also suggests that LLPS can promote and coordinate the process of transcription elongation (Fig. 2)58. In this model, phosphorylation of the C-terminal domain (CTD) of RNA Pol II by CDK7 and CDK9 kinases enables the transition of RNA Pol II from initiation condensates to elongation condensates72–74. These elongation condensates are at least in part composed of the positive transcription elongation factor b (P-TEFb) and various splicing factors73,74. The P-TEFb component cyclin T1 contains an IDR and shows LLPS properties in cells, including fusion events and sensitivity to 1,6-hexanediol73. Specifically, in vitro, the phosphorylated form of the RNA Pol II CTD was shown to preferentially incorporate into SRSF1- and SRSF2-containing splicing factor condensates compared to mediator condensates74. Together, these results highlight the ability of PTMs to rapidly modulate the composition and shuttling of factors between adjacent, yet distinct, biomolecular condensates.
A key consideration when assessing the capacity of macromolecules to undergo LLPS in vivo is the level of expression of the factor of interest. As already stated, biological condensates assemble only after reaching Csat, making them extremely sensitive to changes in protein concentration40. Although exogenous expression of molecular players can reveal their capacity to assemble into biomolecular condensates, caution should be used when employing such an approach. For example, as mentioned above, clear evidence for LLPS was observed upon gross overexpression of FET proteins (FUS and TAF15). However, even if endogenous levels of FET proteins lead to the formation of dynamic hubs that show selectivity for binding partners and RNA Pol II, LLPS features were not observed under these conditions65. This highlights the importance of studying LLPS processes using different systems under physiologically relevant expression levels, as performed in different studies13,62,64,74–76.
LLPS and the spatiotemporal regulation of DNA repair
Cellular health and longevity are continuously threatened by various sources of DNA damage. Therefore, cells have evolved several complex DNA repair pathways. Mounting evidence suggests that different DNA repair pathways rely on LLPS for efficient repair (Fig. 3a)12,13,70,75–77. Among the early responders at sites of DNA damage are members of the ADP-ribosyltransferase (PARP) family78. At the site of DNA damage, PARP-1 synthetizes long negatively charged poly(ADP-ribose) (PAR) chains that are subsequently removed by the PAR glycohydrolase PARG79. It has been proposed that PAR could function as a molecular seed to promote the assembly and LLPS of IDR-containing proteins at the site of DNA damage12. After irradiation-induced DNA damage, members of the FET family rapidly accumulate at DNA damage sites in a PAR-dependent manner80,81. This accumulation is promoted by electrostatic interactions between negatively charged PAR chains and unstructured positively charged arginine-glycine-glycine (RGG) motifs present on FUS and other FET proteins82. The structural flexibility of RGG repeats promotes their multivalent electrostatic interactions with PAR chains to promote LLPS83. Indeed, the accumulation of FET proteins at sites of laser microirradiation is associated with changes in light diffraction, suggesting that the refractive index and mass density at the site of damage differ from those of its surrounding environment upon FET recruitment12. Moreover, FET proteins at the site of DNA damage undergo dynamic exchange with the surrounding nucleoplasm, as revealed by FRAP analysis70. In vitro, activated PARP-1 and FUS lead to the formation of large compartments that are specifically enriched with damaged DNA77. The phase separation properties of FUS may be tightly regulated during the DNA damage response84. For instance, DNA-PK-dependent phosphorylation of FUS prevents its phase separation in vitro33,34. On the other hand, such DNA-PK-driven phosphorylation of FUS can promote its localization to the cytoplasm and aggregation into fibrils84,85. These seemingly contradictory effects of DNA-PK-dependent phosphorylation of FUS on its phase separation behaviour in the nucleus and cytoplasm may reflect, at least in part, the different interactomes of FUS in these subcellular compartments. The transient compartmentalization of damaged DNA by PAR and FUS may contribute to the spatiotemporal regulation and sequential recruitment of DNA repair proteins. For example, the genome caretaker protein 53BP1 appears to be excluded from these PAR/FUS compartments and can only access sites of DNA damage upon PAR and FUS removal12. 53BP1 foci at the sites of DNA damage also exhibit liquid-like properties, including frequent fusion and fission events and sensitivity to 1,6-hexanediol75,76. Of note, the ability of 53BP1 to undergo LLPS depends on its oligomerization domain instead of its unstructured N-terminal domain75. Importantly, this oligomerization domain is also responsible for the multimerization and accumulation of 53BP1 at the site of DNA damage86. In addition, the recruitment and dynamic behaviour of 53BP1 are modulated by the presence of long non-coding RNAs transcribed from the site of DNA damage76,87–89. This local transcription is promoted by MRN-dependent recruitment of the Mediator protein MED1, transcription factors of the preinitiation complex, and RNA Pol II at DNA double strand breaks (DSBs)76. In light of the capacity of MED1 and RNA Pol II to undergo LLPS at super-enhancers, it is possible that these factors also directly promote the assembly of molecular condensates at DNA damage sites.
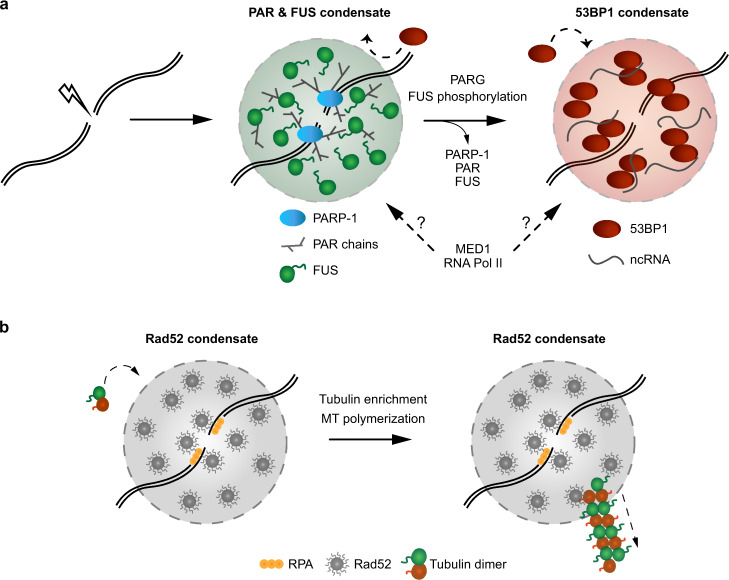
Fig. 3. LLPS in the compartmentalization of damaged DNA.
a Mammalian PARP-1 is an early responder at DNA damage sites and synthetizes long negatively charged PAR chains that interact with positively charged RGG motifs present on FUS proteins. LLPS of FUS allows the compartmentalization of the DNA damage site and the exclusion of specific factors, such as 53BP1. Upon PARG activation and FUS phosphorylation, the FUS condensate disassembles and allows for the enrichment and subsequent LLPS of 53BP1. This phenomenon is mediated, at least in part, by the oligomerization domain of 53BP1. The enrichment of the mediator component MED1 and RNA Pol II in either compartment remains to be determined. b Budding yeast Rad52 forms biomolecular condensates at sites of DNA damage. The condensates can concentrate tubulin and drive the polymerization of microtubule filaments. The filaments help mobilize damaged DNA inside of the nucleus to promote repair.
The role of phase separation in DNA repair appears to be evolutionarily conserved. Work performed with Saccharomyces cerevisiae revealed that the DNA repair and recombination protein Rad52 exhibits liquid-like behaviour in vitro and in vivo13. Surprisingly, sequential fusion of smaller Rad52 droplets inside of the nucleus results in a large liquid-like Rad52 droplet that becomes enriched in tubulin before projecting microtubule filaments from clustered sites of DNA damage (Fig. 3b). These DNA damage-induced intranuclear microtubule filaments (DIMs) contribute to DNA repair by increasing the mobilization of damaged chromatin within the nucleus13,90. The future identification of factors that may be enriched or excluded from such nuclear Rad52 condensates could provide additional unique insights into the relationship between LLPS and DNA repair.
Conclusions and future directions
The growing impact of phase separation across cell and molecular biology has challenged our understanding of intracellular organization. The physicochemical properties of LLPS that underlie the assembly of various biomolecular condensates allow for a wide range of possible functions. Indeed, LLPS can efficiently organize the nucleus into distinct and specialized subcompartments that greatly differ in size and composition. These various condensates can serve to filter for specific biomolecular interactions that can either activate or restrict different biomolecular processes. LLPS is also emerging as a powerful mechanism in the control of biochemical reaction chains. Specifically, this is achieved by segregating, in space and/or time, the subsequent steps of a reaction or pathway into separate biomolecular condensates. These distinct condensates can be physically separated but still allowed to communicate with each other via the transfer of activated factors such as phosphorylated RNA Pol II from one condensate to the other58,74. However, distinct biomolecular condensates may independently assemble and disassemble at specific genomic locations after performing certain functions, such as DNA damage repair12.
The recent advances in the field of LLPS also raise several questions regarding the precise regulation and interplay of distinct biomolecular condensates inside of the nucleus. For example, how are the phase separation properties of heterochromatin affected by DNA damage in this compartment? One possibility is that pre-existing phase-separated compartments (such as an HP1-driven heterochromatin compartment) at sites of DNA damage might first need to disassemble to allow access to DNA repair factors and their organization via LLPS de novo. Consistent with this possibility, the assembly of biomolecular condensates can produce mechanical forces that are capable of dynamically restructuring the genome91. Such forces, combined with the action of chromatin remodelling enzymes, could enable the formation of low-chromatin density areas that favour the recruitment of DNA repair factors. It will be important to determine whether such mechanical reorganizations occur within heterochromatin compartments upon DNA damage. Alternatively, the mobilization of damaged DNA sites away from pre-existing phase-separated compartments might be required before the de novo assembly of biomolecular condensates favouring genome repair at the site of damage. In fact, the relocation of damaged DNA outside of repair-repressive nuclear domains does assist homologous recombination factors in gaining access to damaged DNA92–94. Moreover, in S. cerevisiae, Rad52 liquid-like droplets are assembled at damaged DNA and then transported onto microtubule filaments within the nucleus to promote repair13. Thus, future research should further explore the interplay between LLPS, damaged DNA movement and repair in different organisms95. In-line with these ideas, do the differences in phase separation properties between heterochromatin and euchromatin enable the specific recruitment of repair proteins at DNA damage sites? If different condensates constitute different “chromatin niches” for different DNA repair factors, this may also contribute to or even dictate DNA repair pathway choices and the level of activation of DNA damage checkpoint signalling. More globally, what factors are specifically enriched within or excluded from various biomolecular condensates? The identification of these factors through large-scale proteomics or imaging approaches will enable researchers to better appreciate the complexity and roles of these structures. For example, the identification of P body components paved the way to a better understanding of their regulation and functions in cells96. Finally, can we develop tools to allow us to study the features of individual biomolecular condensates in vivo? Thus, despite the immense global effort to understand the life cycles and functions of biomolecular condensates, when we consider the state of our knowledge of LLPS, it is clear that we still have many more questions than answers. Future studies that aim to answer these questions will undoubtedly provide unique insights into the intricate inner workings of the cell and may shed light on numerous human diseases.
Acknowledgements
We thank Mekhail lab members for helpful discussions. This work was supported by grants to K.M. from the Canadian Institutes of Health Research (299429, 388041) and Canada Research Chairs (950-230661).
Author contributions
G.L. wrote the manuscript and prepared the figures. G.L. and K.M. conceived the topic, conducted revisions, and wrote reviewer rebuttals. K.M. supervised the writing and edited the text and figures.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng A, Weber SC. Evidence for and against liquid-liquid phase separation in the nucleus. Noncoding RNA. 2019;5:50. doi: 10.3390/ncrna5040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 5.Ditlev JA, Case LB, Rosen MK. Who’s in and who’s out-compositional control of biomolecular condensates. J. Mol. Biol. 2018;430:4666–4684. doi: 10.1016/j.jmb.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 7.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson AG, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plys AJ, Kingston RE. Dynamic condensates activate transcription. Science. 2018;361:329–330. doi: 10.1126/science.aau4795. [DOI] [PubMed] [Google Scholar]
- 12.Altmeyer M, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat. Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshidari R, et al. DNA repair by Rad52 liquid droplets. Nat. Commun. 2020;11:695. doi: 10.1038/s41467-020-14546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abraham KJ, et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature. 2020;585:298–302. doi: 10.1038/s41586-020-2497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguzzi A, Altmeyer M. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016;26:547–558. doi: 10.1016/j.tcb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Forman-Kay JD, Kriwacki RW, Seydoux G. Phase separation in biology and disease. J. Mol. Biol. 2018;430:4603–4606. doi: 10.1016/j.jmb.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberti S, Dormann D. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 18.Babinchak WM, Surewicz WK. Liquid-liquid phase separation and its mechanistic role in pathological protein aggregation. J. Mol. Biol. 2020;432:1910–1925. doi: 10.1016/j.jmb.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pak CW, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vernon RM, et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. elife. 2018;7:e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeynaems S, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. elife. 2014;3:e04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fay MM, Anderson PJ. The role of RNA in biological phase separations. J. Mol. Biol. 2018;430:4685–4701. doi: 10.1016/j.jmb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Jove Navarro M, et al. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 2019;10:3230. doi: 10.1038/s41467-019-11241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon Y, Chung YD. RNA-mediated regulation of chromatin structures. Genes Genomics. 2020;42:609–617. doi: 10.1007/s13258-020-00929-5. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017;292:19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 31.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Bah A, Forman-Kay JD. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monahan Z, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e616. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owen I, Shewmaker F. The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int. J. Mol. Sci. 2019;20:5501. doi: 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riback JA, et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040.e1019. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrenberg L. Influence of temperature on the nucleolus and its coacervate nature. Hereditas. 1946;32:407–418. doi: 10.1111/j.1601-5223.1946.tb02783.x. [DOI] [PubMed] [Google Scholar]
- 38.Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feric M, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 41.McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019;33:1619–1634. doi: 10.1101/gad.331520.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Jia ST, Jia S. New insights into the regulation of heterochromatin. Trends Genet. 2016;32:284–294. doi: 10.1016/j.tig.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allshire RC, Madhani HD. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018;19:229–244. doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostrowski LA, et al. Conserved Pbp1/Ataxin-2 regulates retrotransposon activity and connects polyglutamine expansion-driven protein aggregation to lifespan-controlling rDNA repeats. Commun. Biol. 2018;1:187. doi: 10.1038/s42003-018-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/S1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 49.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 50.Brasher SV, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000;10:517–525. doi: 10.1016/S0960-9822(00)00467-X. [DOI] [PubMed] [Google Scholar]
- 52.Machida S, et al. Structural Basis of heterochromatin formation by human HP1. Mol. Cell. 2018;69:385–397.e388. doi: 10.1016/j.molcel.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Sanulli S, et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature. 2019;575:390–394. doi: 10.1038/s41586-019-1669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erdel F, et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell. 2020;78:236–249.e237. doi: 10.1016/j.molcel.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson BA, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.e421. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017;18:246–262. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res. 2020;30:393–407. doi: 10.1038/s41422-020-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 59.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabari BR, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boija A, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855.e1816. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho WK, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong S, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai D, et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019;21:1578–1589. doi: 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu Y, et al. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020;22:453–464. doi: 10.1038/s41556-020-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han X, et al. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat. Struct. Mol. Biol. 2020;27:333–341. doi: 10.1038/s41594-020-0394-8. [DOI] [PubMed] [Google Scholar]
- 69.Kwon I, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 71.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boehning M, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018;25:833–840. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- 73.Lu H, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318–323. doi: 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kilic S, et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pessina F, et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019;21:1286–1299. doi: 10.1038/s41556-019-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singatulina AS, et al. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019;27:1809–1821.e1805. doi: 10.1016/j.celrep.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 78.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jungmichel S, et al. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 80.Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rulten SL, et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 83.Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44:993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deng Q, et al. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 2014;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Naumann M, et al. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat. Commun. 2018;9:335. doi: 10.1038/s41467-017-02299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mirman Z, de Lange T. 53BP1: a DSB escort. Genes Dev. 2020;34:7–23. doi: 10.1101/gad.333237.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pryde F, et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- 88.Francia S, Cabrini M, Matti V, Oldani A, d’Adda di Fagagna F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 2016;129:1468–1476. doi: 10.1242/jcs.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Michelini F, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oshidari R, et al. Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat. Commun. 2018;9:2567. doi: 10.1038/s41467-018-05009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin Y, et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175:1481–1491. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Torres-Rosell J, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 93.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ryu T, et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oshidari R, Mekhail K, Seeber A. Mobility and repair of damaged DNA: random or directed? Trends Cell Biol. 2019;30:144–156. doi: 10.1016/j.tcb.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 96.Hubstenberger A, et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017;68:144–157.e145. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]