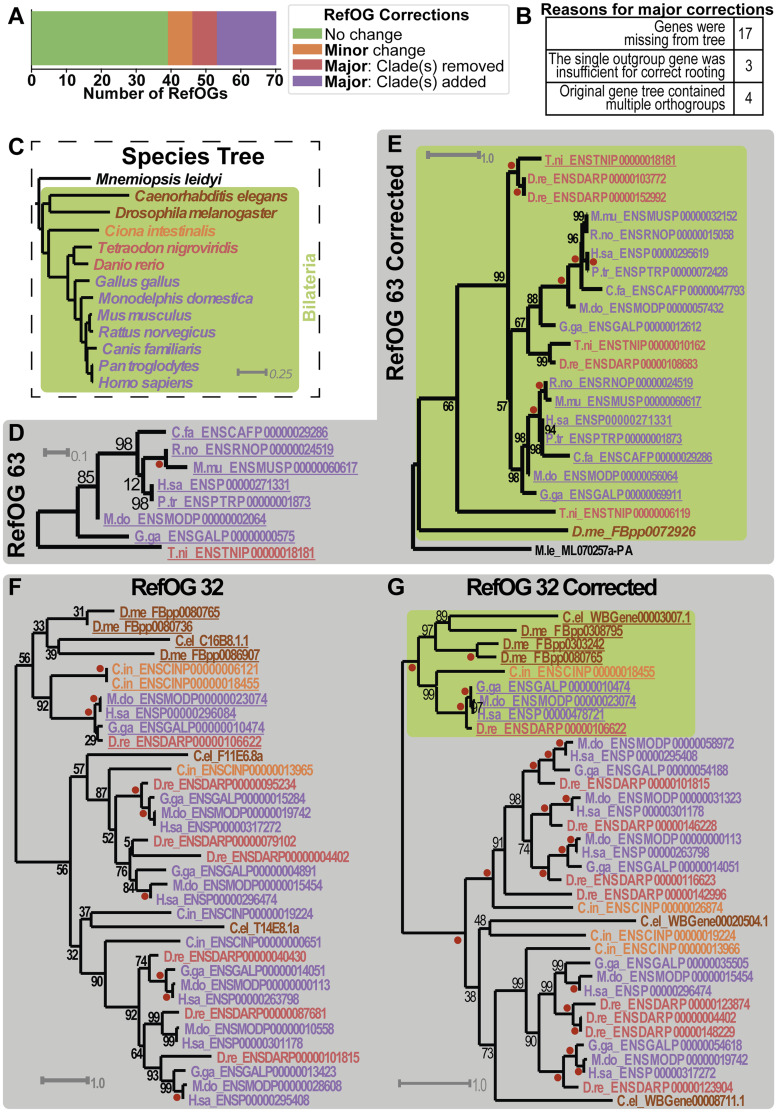
Fig. 1.
Evaluation and revision of RefOGs from Orthobench. (A) Summary of the corrections made to the RefOG data set. (B) Reasons for major corrections to RefOGs from the previous study. (C) The species tree. Green shaded area shows the 12 Bilaterian species for which the Bilateria-level orthogroups (RefOGs) were defined. One outgroup species, which appears in the gene trees in the figure, is also shown. (D) Example of a major improvement for which clades had been missing from the original RefOG tree: RefOG 63 gene tree as determined in the original study. (E) Gene tree from this study showing the corrected RefOG 63 orthogroup shaded green. Phylogenetic analysis revealed that the original RefOG32 comprises two separate orthogroups that diverged at a gene duplication even preceding the divergence of the vertebrates. (F) Example of a major improvement for which extra clades of genes had been included in the original RefOG: RefOG 32 gene tree as determined in the original study. (G) Gene tree from this study showing the corrected RefOG 32 orthogroup. Phylogenetic analysis revealed that these genes diverged from the remaining genes in the tree at a gene duplication event predating the divergence of the Deuterostomes and Protostomes. Gene trees show previously identified orthogroup containing the newly delimited orthogroup from this study (green shaded clade). Genes/species are colored according to species. Corresponding genes identified as members of the orthogroup in both studies are underlined (including when identifiers have been updated). Red dot = 100% bootstrap support.