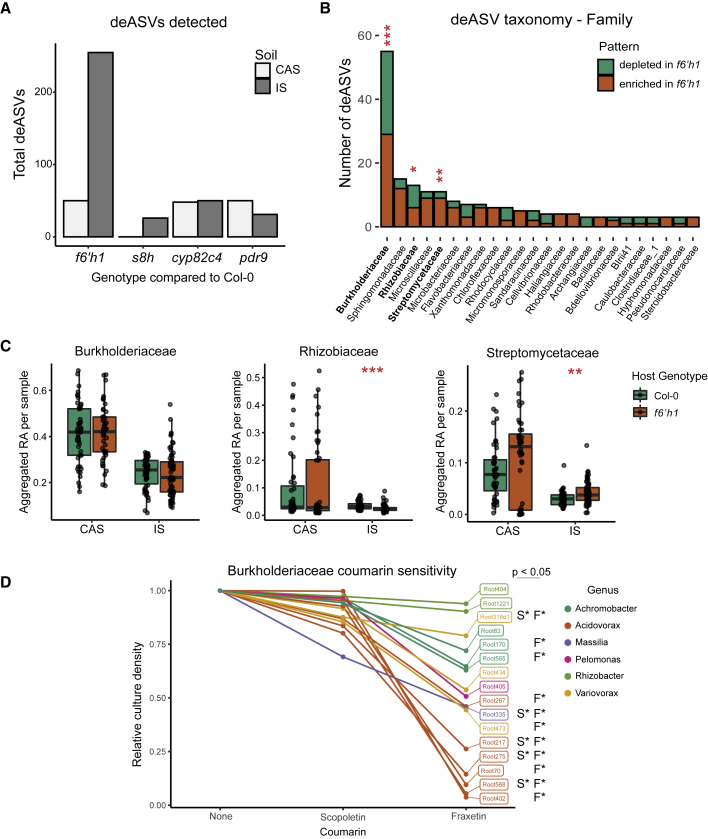
Figure 2.
Coumarin Biosynthesis Restructures the Root Microbiota at the ASV Level
(A) Number of deASVs detected in indicated mutants compared with Col-0 in each soil. Data are pooled from three experiments (except s8h, which was included in only one), and filtered for ASVs found in at least three samples with RA > 0.05%. Differential enrichment was calculated using a negative binomial generalized log-linear model at an FDR-adjusted p value of 0.05.
(B) Family-level taxonomic classification of deASVs in f6’h1 plants growing on IS. Colors indicate if deASVs were enriched or depleted in f6’h1 compared with Col-0. Hypergeometric enrichment test was performed to determine if each family was over- or under-represented in deASV list compared with all detected ASVs. Red asterisks indicate significance with FDR-adjusted p values.
(C) Sample-wise aggregated relative abundance of the top three families most significantly over-represented in deASVs: Burkholderiaceae, Rhizobiaceae, and Streptomycetaceae. Each data point represents the average RA aggregated at the family level in a single sample. Significance between genotypes in each soil was determined by Wilcoxon ranked sum test.
(D) Overnight growth of Burkholderiaceae bacterial strains in the presence of scopoletin or fraxetin. Optical density (OD) of cultures was normalized to the OD of each strain in the absence of coumarins. Significant differences (p ≤ 0.05 by Tukey’s HSD) in growth compared with the control are indicated for scopoletin (S∗) and fraxetin (F∗) to the right of each strain. Data are averages of 2–4 experiments, each with 2–3 technical replicates, per strain. ∗, ∗∗, and ∗∗∗ in (B) and (C) indicate p ≤ 0.05, 0.01, and 0.001, respectively. See also Figures S2 and S3.