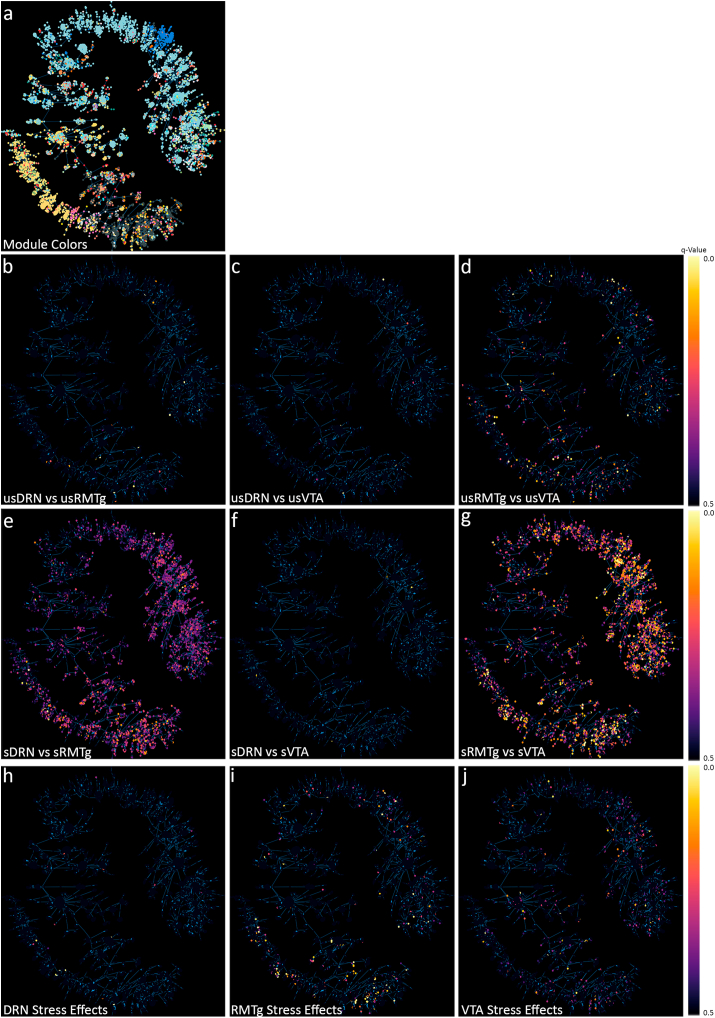
Fig. 5.
Minimum spanning trees of WGCNA results. a) Two-dimensional minimum spanning trees illustrate the 35 gene modules identified by weighted gene coexpression network analysis (WGCNA). Using the original DESeq2 analysis, the q-values for each individual gene is shown for pairwise comparisons of unstressed DRN to unstressed RMTg (b), unstressed DRN to unstressed VTA (c), and unstressed RMTg to unstressed VTA (d). There were relatively few changes apparent between the pathways in unstressed animals, but after stress, there were larger differences apparent in in the stressed DRN vs. stressed RMTg (e), stressed DRN vs. stressed VTA (f), and stressed RMTg vs. stressed VTA (g). The RMTg vs. VTA showed the most significant differences. (h–j) shows the pairwise comparisons for stressed vs. unstressed DRN (h), RMTg (i), and VTA (j).